
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,531,096 – 21,531,187 |
| Length | 91 |
| Max. P | 0.700588 |

| Location | 21,531,096 – 21,531,187 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 65.20 |
| Shannon entropy | 0.70323 |
| G+C content | 0.62779 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -11.65 |
| Energy contribution | -13.01 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
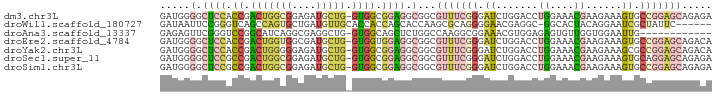
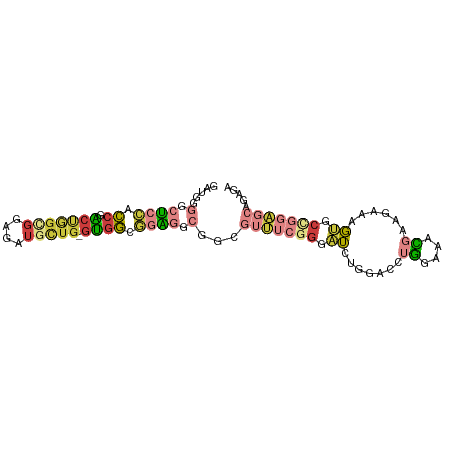
>dm3.chr3L 21531096 91 + 24543557 GAUGGGGCUCCACCGACUGGCGGAGAUGCUG-GUGGCGGAGGCGGCGUUUCGGGAUCUGGACCUGGAAACGAAGAAAGUGCCGGAGCAGAGA ......(((((.(((((..(((....)))..-))..))).((((.((((((.((........)).)))))).......)))))))))..... ( -33.70, z-score = -1.01, R) >droWil1.scaffold_180727 1612065 85 - 2741493 GAUAAUUCCGGGUCAGCCAGUGCUGAUGUUGCACCACCAGCACCAAGCGCAGGGGAACGAGGC-GGCACUACAGGAAUCGCUAUUC------ ....(((((.((....))(((((((.(.(((..((.((.((.......)).))))..))).))-))))))...)))))........------ ( -24.90, z-score = -0.20, R) >droAna3.scaffold_13337 6190567 79 - 23293914 GAGAGUUCGGGUCCGGCAUCAGGCGAGGCUG-GUGGCAGCUCUGGCCAAGGCGGAAACGUGGAGAGUGUUGGUGGAAUUG------------ ...(((((.(..((((((((.(((.((((((-....)))).)).)))...(((....)))...)).))))))).))))).------------ ( -30.30, z-score = -2.10, R) >droEre2.scaffold_4784 21184407 91 + 25762168 GAUGGGGCUCCACCGACUGGUGGCGAUGCUG-GUGGUGGAGGCGGCGUUUCGGGAUCUGGACCUGGAAACGAAGAAAGUGCCGGAGCAGACA .....(.(((((((.((..((......))..-))))))))).)...(((((((.(.((.....((....)).....))).)))))))..... ( -35.20, z-score = -1.46, R) >droYak2.chr3L 5212417 91 - 24197627 GAUGGGGCUCCACCGACUGGGGGAGAUGCUG-GUGGCGGAGGCGGCGUUUCGGGAUCUGGACCUGGAAACGAAGAAAGCGCCGGAGCAGACA ..((..(((((.(((((..(.(....).)..-))..))).((((.((((((.((........)).)))))).......)))))))))...)) ( -31.70, z-score = -0.22, R) >droSec1.super_11 1450896 91 + 2888827 GAUGGGGCUCCGCCGACUGGCGGAGAUGCUG-GUGGCGGAGGCGGCGUUUCGGGAUCUGGACCUGGAAACGAAGAAAGUGCAGGAGCAGAGA .....(.(((((((.((..(((....)))..-))))))))).)..((((((.((........)).)))))).......(((....))).... ( -36.50, z-score = -2.14, R) >droSim1.chr3L 20864398 91 + 22553184 GAUGGGGCUCCGCCGACUGGCGGAGAUGCUG-GUGGCGGAGGCGGCGUUUCGGGAUCUGGACCUGGAAACGAAGAAAGUGCCGGAGCAGAGA .....(.(((((((.((..(((....)))..-))))))))).)...(((((((.(.((.....((....)).....))).)))))))..... ( -39.00, z-score = -2.39, R) >consensus GAUGGGGCUCCACCGACUGGCGGAGAUGCUG_GUGGCGGAGGCGGCGUUUCGGGAUCUGGACCUGGAAACGAAGAAAGUGCCGGAGCAGAGA .......(((((((....)))))))..(((....))).........(((((((.((.......((....))......)).)))))))..... (-11.65 = -13.01 + 1.36)
| Location | 21,531,096 – 21,531,187 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 65.20 |
| Shannon entropy | 0.70323 |
| G+C content | 0.62779 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -7.23 |
| Energy contribution | -8.99 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21531096 91 - 24543557 UCUCUGCUCCGGCACUUUCUUCGUUUCCAGGUCCAGAUCCCGAAACGCCGCCUCCGCCAC-CAGCAUCUCCGCCAGUCGGUGGAGCCCCAUC ....((((..(((........((((((..(((....)))..))))))........)))..-.)))).(((((((....)))))))....... ( -25.79, z-score = -1.77, R) >droWil1.scaffold_180727 1612065 85 + 2741493 ------GAAUAGCGAUUCCUGUAGUGCC-GCCUCGUUCCCCUGCGCUUGGUGCUGGUGGUGCAACAUCAGCACUGGCUGACCCGGAAUUAUC ------.......((((((.((...(((-....(((......)))...((((((((((......)))))))))))))..))..))))))... ( -29.30, z-score = -1.37, R) >droAna3.scaffold_13337 6190567 79 + 23293914 ------------CAAUUCCACCAACACUCUCCACGUUUCCGCCUUGGCCAGAGCUGCCAC-CAGCCUCGCCUGAUGCCGGACCCGAACUCUC ------------.....................((..((((.(..(((.((.((((....-)))))).)))....).))))..))....... ( -19.40, z-score = -2.24, R) >droEre2.scaffold_4784 21184407 91 - 25762168 UGUCUGCUCCGGCACUUUCUUCGUUUCCAGGUCCAGAUCCCGAAACGCCGCCUCCACCAC-CAGCAUCGCCACCAGUCGGUGGAGCCCCAUC .....((((((((........((((((..(((....)))..))))))..((.........-..))...)))(((....))))))))...... ( -23.30, z-score = -1.01, R) >droYak2.chr3L 5212417 91 + 24197627 UGUCUGCUCCGGCGCUUUCUUCGUUUCCAGGUCCAGAUCCCGAAACGCCGCCUCCGCCAC-CAGCAUCUCCCCCAGUCGGUGGAGCCCCAUC .....(((((((((.......((((((..(((....)))..)))))).......))))((-(.((..........)).))))))))...... ( -25.64, z-score = -1.43, R) >droSec1.super_11 1450896 91 - 2888827 UCUCUGCUCCUGCACUUUCUUCGUUUCCAGGUCCAGAUCCCGAAACGCCGCCUCCGCCAC-CAGCAUCUCCGCCAGUCGGCGGAGCCCCAUC ..((((..((((.((.......))...))))..))))..............(((((((((-..((......))..)).)))))))....... ( -24.90, z-score = -1.99, R) >droSim1.chr3L 20864398 91 - 22553184 UCUCUGCUCCGGCACUUUCUUCGUUUCCAGGUCCAGAUCCCGAAACGCCGCCUCCGCCAC-CAGCAUCUCCGCCAGUCGGCGGAGCCCCAUC ....((((..(((........((((((..(((....)))..))))))........)))..-.)))).(((((((....)))))))....... ( -27.69, z-score = -2.34, R) >consensus UCUCUGCUCCGGCACUUUCUUCGUUUCCAGGUCCAGAUCCCGAAACGCCGCCUCCGCCAC_CAGCAUCUCCGCCAGUCGGCGGAGCCCCAUC .....................((((((..(((....)))..))))))....(((((((....................)))))))....... ( -7.23 = -8.99 + 1.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:22 2011