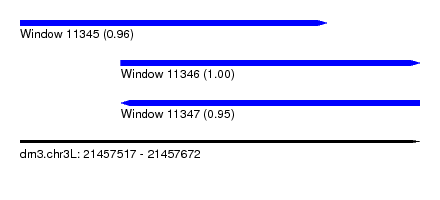
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,457,517 – 21,457,672 |
| Length | 155 |
| Max. P | 0.998162 |

| Location | 21,457,517 – 21,457,636 |
|---|---|
| Length | 119 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.69 |
| Shannon entropy | 0.28024 |
| G+C content | 0.35689 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -18.28 |
| Energy contribution | -17.96 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21457517 119 + 24543557 AGACUCGUUCUCCUCGUUCUCCUCGCUCGCCUCGCUGCAGUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAGGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUA ........................((.......))((((.(((......)))))))..((((...(((((((((((((((.(((....)))))))).))))))))))...))))..... ( -21.30, z-score = -1.70, R) >droMoj3.scaffold_6680 19137804 106 - 24764193 -----------CGUCGUCGUCUUCUUCGUUG--GCUGCAGUUGUUUUCACAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGAUUGUUGUCUCUGAAAAUAUGUUGAUAAAAUA -----------.((((.((.......)).))--))((((.((((....))))))))..((((...(((((((((((((((.((......)))))))).)))))))))...))))..... ( -23.20, z-score = -2.62, R) >droPer1.super_67 19926 108 + 321716 -----------CGGCGUGGUCUCCGUCGCUCCCGCUGCAGUUGUUUUUCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCCUGUUGUCUCUGAAAAUAUGUUGAUAAAACA -----------(((((.((...........)))))))....((((((..((((............(((((((((((((((.((......)))))))).)))))))))))))..)))))) ( -24.30, z-score = -2.37, R) >dp4.chrXR_group5 30442 108 - 740492 -----------CGGCGUGGUCUCCGUCGCUCCCGCUGCAGUUGUUUUUCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCCUGUUGUCUCUGAAAAUAUGUUGAUAAAACA -----------(((((.((...........)))))))....((((((..((((............(((((((((((((((.((......)))))))).)))))))))))))..)))))) ( -24.30, z-score = -2.37, R) >droAna3.scaffold_13337 19899573 110 + 23293914 ---------AGACUUUUCUCGUUCUUCCCAACCGUUGCAGUUGUUUUCCCAGUGUAAAUAUUUAAAUAUUUUCAAGACAAACACCGGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUA ---------...................((((..(((((.(((......))))))))........(((((((((((((((.(((....))))))))).)))))))))))))........ ( -21.70, z-score = -2.26, R) >droEre2.scaffold_4784 21115961 110 + 25762168 ---------AGACUCGUUCUCCUCGCUCGCCUCGCUGCAGUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUA ---------.((..((..(.....)..))..))..((((.(((......)))))))..((((...(((((((((((((((.(((....))))))))).)))))))))...))))..... ( -19.10, z-score = -1.54, R) >droYak2.chr3L 5134578 110 - 24197627 ---------AGACUCGUUCUCCUCGCUCGCCUCGCUGCAGUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUA ---------.((..((..(.....)..))..))..((((.(((......)))))))..((((...(((((((((((((((.(((....))))))))).)))))))))...))))..... ( -19.10, z-score = -1.54, R) >droSec1.super_11 1381040 110 + 2888827 ---------AGACUCGUUCUCCUCGCUCGCCUCGCUGCAGUUGUUUUCUCAAUGUAAAUAUUUAAAUAUUUUCAGGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUA ---------.((..((..(.....)..))..))..((((.(((......)))))))..((((...(((((((((((((((.(((....)))))))).))))))))))...))))..... ( -22.00, z-score = -1.88, R) >droSim1.chr3L 20794025 110 + 22553184 ---------AGACUCGUUCUCCUCGCUCGCCUCGCUGCAGUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAGGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUA ---------.((..((..(.....)..))..))..((((.(((......)))))))..((((...(((((((((((((((.(((....)))))))).))))))))))...))))..... ( -21.50, z-score = -1.86, R) >consensus _________AGACUCGUUCUCCUCGCUCGCCUCGCUGCAGUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUA ...................................((((.(((......)))))))..((((...(((((((((((((((.((......)))))))).)))))))))...))))..... (-18.28 = -17.96 + -0.32)

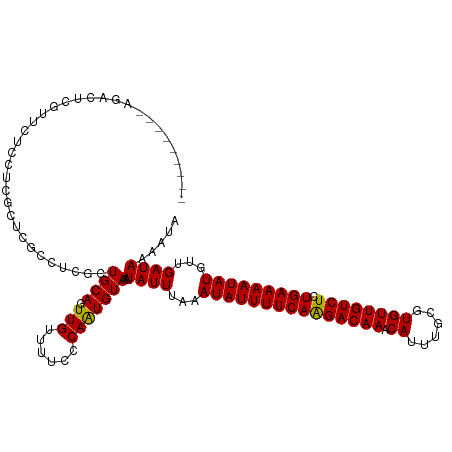
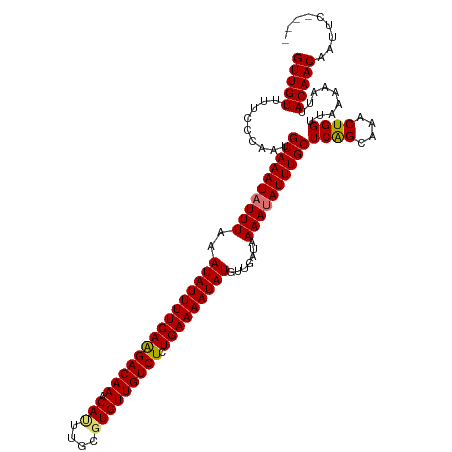
| Location | 21,457,556 – 21,457,672 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.84 |
| Shannon entropy | 0.10077 |
| G+C content | 0.28096 |
| Mean single sequence MFE | -26.99 |
| Consensus MFE | -25.34 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21457556 116 + 24543557 GUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAGGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUAUUUGCUCAGCAAACUGGUUAAAAAUUACAACAAUUC---- (((((.........((((((((((..(((((((((((((((.(((....)))))))).)))))))))).......))))))))))((((....)))).........))))).....---- ( -28.60, z-score = -4.09, R) >droPer1.super_67 19954 120 + 321716 GUUGUUUUUCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCCUGUUGUCUCUGAAAAUAUGUUGAUAAAACAUUUGCUCGGUAAACUGGUUAAAAAUUACAACAAUUCCCCG (((((..((....(((...((((...(((((((((((((((.((......)))))))).)))))))))...))))..)))((((..(((....)))..))))))..)))))......... ( -23.60, z-score = -2.83, R) >dp4.chrXR_group5 30470 120 - 740492 GUUGUUUUUCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCCUGUUGUCUCUGAAAAUAUGUUGAUAAAACAUUUGCUCGGUAAACUGGUUAAAAAUUACAACAAUUCCCCG (((((..((....(((...((((...(((((((((((((((.((......)))))))).)))))))))...))))..)))((((..(((....)))..))))))..)))))......... ( -23.60, z-score = -2.83, R) >droAna3.scaffold_13337 19899603 119 + 23293914 GUUGUUUUCCCAGUGUAAAUAUUUAAAUAUUUUCAAGACAAACACCGGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUAUUUGCUCAGCAAACUGGUUAAAAAUUACAACAAUUCACG- (((((((..(((((((((((((((..(((((((((((((((.(((....))))))))).))))))))).......)))))))))).......)))))..)).....)))))........- ( -30.51, z-score = -4.34, R) >droEre2.scaffold_4784 21115991 116 + 25762168 GUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUAUUUGCUCAGCAAACUGGUUAAAAAUUACAACAAUUC---- (((((.........((((((((((..(((((((((((((((.(((....))))))))).))))))))).......))))))))))((((....)))).........))))).....---- ( -26.20, z-score = -3.56, R) >droYak2.chr3L 5134608 116 - 24197627 GUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUAUUUGCUCAGCAAACUGGUUAAAAAUUACAACAAUUC---- (((((.........((((((((((..(((((((((((((((.(((....))))))))).))))))))).......))))))))))((((....)))).........))))).....---- ( -26.20, z-score = -3.56, R) >droSec1.super_11 1381070 116 + 2888827 GUUGUUUUCUCAAUGUAAAUAUUUAAAUAUUUUCAGGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUAUUUGCUCAGCAAACUGGUUAAAAAUUACAACAAUUC---- (((((.........((((((((((..(((((((((((((((.(((....)))))))).)))))))))).......))))))))))((((....)))).........))))).....---- ( -28.60, z-score = -4.25, R) >droSim1.chr3L 20794055 116 + 22553184 GUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAGGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUAUUUGCUCAGCAAACUGGUUAAAAAUUACAACAAUUC---- (((((.........((((((((((..(((((((((((((((.(((....)))))))).)))))))))).......))))))))))((((....)))).........))))).....---- ( -28.60, z-score = -4.09, R) >consensus GUUGUUUUCCCAAUGUAAAUAUUUAAAUAUUUUCAAGACAAACAUUUGCGUGUUGUCUCUGAAAAUAUGUUGAUAAAAUAUUUGCUCAGCAAACUGGUUAAAAAUUACAACAAUUC____ (((((.........((((((((((..(((((((((((((((.(((....))))))))).))))))))).......))))))))))((((....)))).........)))))......... (-25.34 = -25.34 + 0.00)
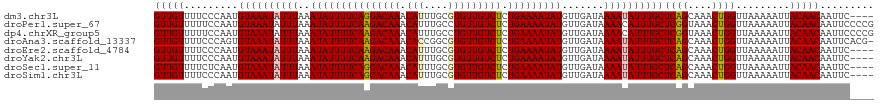
| Location | 21,457,556 – 21,457,672 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.84 |
| Shannon entropy | 0.10077 |
| G+C content | 0.28096 |
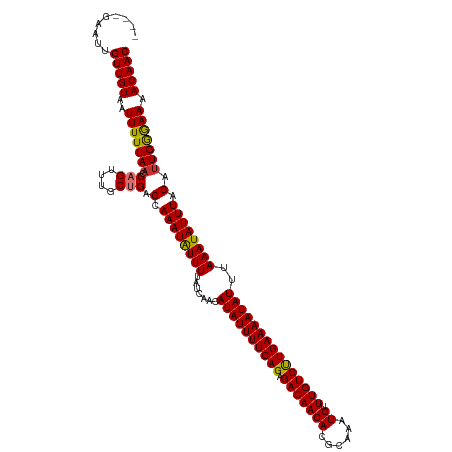
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -21.85 |
| Energy contribution | -21.51 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21457556 116 - 24543557 ----GAAUUGUUGUAAUUUUUAACCAGUUUGCUGAGCAAAUAUUUUAUCAACAUAUUUUCAGAGACAACACGCAAAUGUUUGUCCUGAAAAUAUUUAAAUAUUUACAUUGGGAAAACAAC ----.....(((((..(((((((.(((....))).(.((((((((.......((((((((((.(((((((......)).)))))))))))))))..)))))))).).))))))).))))) ( -23.90, z-score = -2.45, R) >droPer1.super_67 19954 120 - 321716 CGGGGAAUUGUUGUAAUUUUUAACCAGUUUACCGAGCAAAUGUUUUAUCAACAUAUUUUCAGAGACAACAGGCAAAUGUUUGUCUUGAAAAUAUUUAAAUAUUUACAUUGGAAAAACAAC (((..((((((((.......))).)))))..)))....(((((..(((....(((((((((.((((((((......)).)))))))))))))))....)))...)))))........... ( -25.10, z-score = -2.19, R) >dp4.chrXR_group5 30470 120 + 740492 CGGGGAAUUGUUGUAAUUUUUAACCAGUUUACCGAGCAAAUGUUUUAUCAACAUAUUUUCAGAGACAACAGGCAAAUGUUUGUCUUGAAAAUAUUUAAAUAUUUACAUUGGAAAAACAAC (((..((((((((.......))).)))))..)))....(((((..(((....(((((((((.((((((((......)).)))))))))))))))....)))...)))))........... ( -25.10, z-score = -2.19, R) >droAna3.scaffold_13337 19899603 119 - 23293914 -CGUGAAUUGUUGUAAUUUUUAACCAGUUUGCUGAGCAAAUAUUUUAUCAACAUAUUUUCAGAGACAACACGCCGGUGUUUGUCUUGAAAAUAUUUAAAUAUUUACACUGGGAAAACAAC -........(((((.....((..(((((..((...))((((((((.......(((((((((.(((((((((....))).)))))))))))))))..))))))))..)))))..))))))) ( -25.80, z-score = -1.97, R) >droEre2.scaffold_4784 21115991 116 - 25762168 ----GAAUUGUUGUAAUUUUUAACCAGUUUGCUGAGCAAAUAUUUUAUCAACAUAUUUUCAGAGACAACACGCAAAUGUUUGUCUUGAAAAUAUUUAAAUAUUUACAUUGGGAAAACAAC ----.....(((((..(((((((.(((....))).(.((((((((.......(((((((((.((((((((......)).)))))))))))))))..)))))))).).))))))).))))) ( -21.50, z-score = -1.61, R) >droYak2.chr3L 5134608 116 + 24197627 ----GAAUUGUUGUAAUUUUUAACCAGUUUGCUGAGCAAAUAUUUUAUCAACAUAUUUUCAGAGACAACACGCAAAUGUUUGUCUUGAAAAUAUUUAAAUAUUUACAUUGGGAAAACAAC ----.....(((((..(((((((.(((....))).(.((((((((.......(((((((((.((((((((......)).)))))))))))))))..)))))))).).))))))).))))) ( -21.50, z-score = -1.61, R) >droSec1.super_11 1381070 116 - 2888827 ----GAAUUGUUGUAAUUUUUAACCAGUUUGCUGAGCAAAUAUUUUAUCAACAUAUUUUCAGAGACAACACGCAAAUGUUUGUCCUGAAAAUAUUUAAAUAUUUACAUUGAGAAAACAAC ----.....(((((..(((((((.(((....))).(.((((((((.......((((((((((.(((((((......)).)))))))))))))))..)))))))).).))))))).))))) ( -24.80, z-score = -3.13, R) >droSim1.chr3L 20794055 116 - 22553184 ----GAAUUGUUGUAAUUUUUAACCAGUUUGCUGAGCAAAUAUUUUAUCAACAUAUUUUCAGAGACAACACGCAAAUGUUUGUCCUGAAAAUAUUUAAAUAUUUACAUUGGGAAAACAAC ----.....(((((..(((((((.(((....))).(.((((((((.......((((((((((.(((((((......)).)))))))))))))))..)))))))).).))))))).))))) ( -23.90, z-score = -2.45, R) >consensus ____GAAUUGUUGUAAUUUUUAACCAGUUUGCUGAGCAAAUAUUUUAUCAACAUAUUUUCAGAGACAACACGCAAAUGUUUGUCUUGAAAAUAUUUAAAUAUUUACAUUGGGAAAACAAC .........(((((..(((((((.(((....))).(.((((((((.......((((((((((.(((((((......)).)))))))))))))))..)))))))).).))))))).))))) (-21.85 = -21.51 + -0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:15 2011