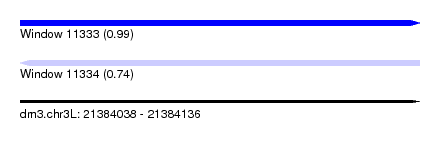
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,384,038 – 21,384,136 |
| Length | 98 |
| Max. P | 0.986278 |

| Location | 21,384,038 – 21,384,136 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 57.32 |
| Shannon entropy | 0.73848 |
| G+C content | 0.27045 |
| Mean single sequence MFE | -14.92 |
| Consensus MFE | -5.04 |
| Energy contribution | -5.40 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
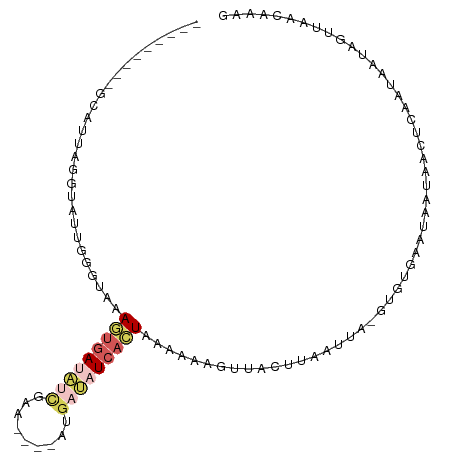
>dm3.chr3L 21384038 98 + 24543557 CUUUUUAAUGUAUUAUCGAGCUAUUGUUCACAC-UAAAUAACUAACAUUUUUAGUGAUAUCAU----UUCGAUAUCACUUUACCCAAUACCUAAUGCUAUAAA--- .........((((((..((((....))))....-..................(((((((((..----...)))))))))............))))))......--- ( -17.90, z-score = -4.28, R) >droAna3.scaffold_13337 10200688 83 + 23293914 ---UAUCUGAAAAUCUUAUCUUUUUGUGAGUGUGUAGUUAAGGGAAAACUUUAAAGAAUAUCU-----UGUAAGUUAUUUAUCACAUUUCA--------------- ---.....((((..(((((......)))))((((...((((((.....)))))).(((((.((-----....)).)))))..)))))))).--------------- ( -9.80, z-score = 0.70, R) >droEre2.scaffold_4784 21041791 93 + 25762168 ------------UUACAGAAUUAUUAUUCACAC-UAAUUAAAUAACUUUUUUAGUGAGUGCGUGUACUCGAAUAUCAUUUUUCCCAUUACCAAUUAUGGCUAUGAA ------------....(((((...(((((..((-(((..((.....))..)))))(((((....))))))))))..)))))...(((..(((....)))..))).. ( -14.90, z-score = -1.67, R) >droSec1.super_11 1308279 92 + 2888827 CUUUGCUAACUAUUAUUGAGUUAUUAUUCACAC-UAAUUAAGUAACUUUUUUAGUGAUAUCAU----UUCGAUAUCACUUGACCCAAUACAUAAUGC--------- .............(((((.((((........((-(.....))).........(((((((((..----...))))))))))))).)))))........--------- ( -15.60, z-score = -2.65, R) >droSim1.chr3L 20717879 92 + 22553184 CUUUGCUAACUAUUAUUGAGUUAUUAUUCACAC-UAAUUAAGUAACUAUUUUAGUGAUAUCAU----UUCGAUAUCACUUGACCCAAUACCUAAAGC--------- ....(((......(((((.((((........((-(.....))).........(((((((((..----...))))))))))))).))))).....)))--------- ( -16.40, z-score = -3.02, R) >consensus CUUUGCUAACUAUUAUUGAGUUAUUAUUCACAC_UAAUUAAGUAACUUUUUUAGUGAUAUCAU____UUCGAUAUCACUUGACCCAAUACCUAAUGC_________ .................((((....)))).......................(((((((((.........)))))))))........................... ( -5.04 = -5.40 + 0.36)


| Location | 21,384,038 – 21,384,136 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 57.32 |
| Shannon entropy | 0.73848 |
| G+C content | 0.27045 |
| Mean single sequence MFE | -16.66 |
| Consensus MFE | -4.36 |
| Energy contribution | -5.52 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.744206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21384038 98 - 24543557 ---UUUAUAGCAUUAGGUAUUGGGUAAAGUGAUAUCGAA----AUGAUAUCACUAAAAAUGUUAGUUAUUUA-GUGUGAACAAUAGCUCGAUAAUACAUUAAAAAG ---............(.((((((((..(((((((((...----..))))))))).....((((..((((...-..))))..))))))))))))...)......... ( -19.80, z-score = -2.49, R) >droAna3.scaffold_13337 10200688 83 - 23293914 ---------------UGAAAUGUGAUAAAUAACUUACA-----AGAUAUUCUUUAAAGUUUUCCCUUAACUACACACUCACAAAAAGAUAAGAUUUUCAGAUA--- ---------------.....(((((............(-----((.....)))...((((.......))))......))))).....................--- ( -5.00, z-score = 1.20, R) >droEre2.scaffold_4784 21041791 93 - 25762168 UUCAUAGCCAUAAUUGGUAAUGGGAAAAAUGAUAUUCGAGUACACGCACUCACUAAAAAAGUUAUUUAAUUA-GUGUGAAUAAUAAUUCUGUAA------------ (((((.((((....)))).)))))........(((((((((......))))(((((.............)))-))..)))))............------------ ( -14.72, z-score = -0.68, R) >droSec1.super_11 1308279 92 - 2888827 ---------GCAUUAUGUAUUGGGUCAAGUGAUAUCGAA----AUGAUAUCACUAAAAAAGUUACUUAAUUA-GUGUGAAUAAUAACUCAAUAAUAGUUAGCAAAG ---------(((((((.((((((((..(((((((((...----..)))))))))........((((.....)-))).........)))))))))))))..)).... ( -21.30, z-score = -3.62, R) >droSim1.chr3L 20717879 92 - 22553184 ---------GCUUUAGGUAUUGGGUCAAGUGAUAUCGAA----AUGAUAUCACUAAAAUAGUUACUUAAUUA-GUGUGAAUAAUAACUCAAUAAUAGUUAGCAAAG ---------(((.....((((((((..(((((((((...----..)))))))))........((((.....)-))).........))))))))......))).... ( -22.50, z-score = -3.73, R) >consensus _________GCAUUAGGUAUUGGGUAAAGUGAUAUCGAA____AUGAUAUCACUAAAAAAGUUACUUAAUUA_GUGUGAAUAAUAACUCAAUAAUAGUUAACAAAG ...........................((((((((((.......)))))))))).................................................... ( -4.36 = -5.52 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:04 2011