| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,350,196 – 21,350,297 |
| Length | 101 |
| Max. P | 0.939410 |

| Location | 21,350,196 – 21,350,297 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 63.62 |
| Shannon entropy | 0.72965 |
| G+C content | 0.46978 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -8.68 |
| Energy contribution | -10.64 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
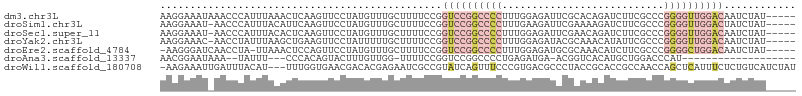
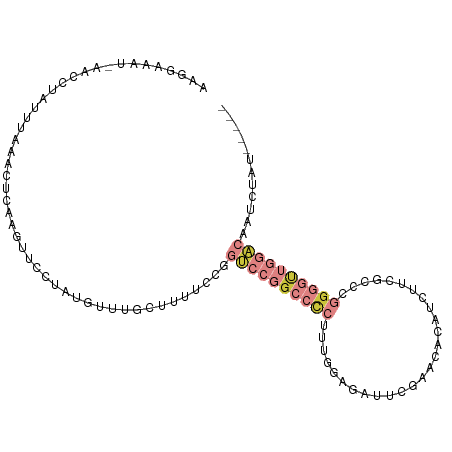
>dm3.chr3L 21350196 101 - 24543557 AAGGAAAUAAACCCAUUUAAACUCAAGUUCCUAUGUUUGCUUUUCCGGUCCGGCCCCUUUGGAGAUUCGCACAGAUCUUCGCCCGGGGUUGGACAAUCUAU----- ..(((((...(..(((...(((....)))...)))..)...))))).((((((((((..((((((((......))))))))...)))))))))).......----- ( -33.70, z-score = -2.87, R) >droSim1.chr3L 20697071 100 - 22553184 AAGGAAAU-AACCCAUUUACAUUCAAGUUCCUAUGUUUGCUUUUCCGGUCCGGCCCCUUUGAAGAUUCGAAAAGAUCUUCGCCCGGGGUUGGACUAUCUAU----- ..(((((.-.(..(((..((......))....)))..)...)))))(((((((((((..((((((((......))))))))...)))))))))))......----- ( -33.60, z-score = -3.28, R) >droSec1.super_11 1287491 100 - 2888827 AAGGAAAU-AACCCAUUUACACUCAAGUUCCUAUGUUUGCUUUUCCGGUCCGGCCCCUUUGGAGAUUCGAACAGAUCUUCGCCCGGGGUUGGACAAUCUAU----- ..(((((.-.(..(((..((......))....)))..)...))))).((((((((((..((((((((......))))))))...)))))))))).......----- ( -32.20, z-score = -2.37, R) >droYak2.chr3L 5037245 100 + 24197627 AAGGAAAC-AACCUAUUUAAGCUGAAGUUCCUAUUUUUGCUUUUCCGGUCCGGCCCCUUUGGAGAUACGCAAACAUAUUCGCCCGGGGUUGGACAAUCUAU----- ..(((((.-..........(((.(((((....))))).)))))))).((((((((((...((.((.............)).)).)))))))))).......----- ( -28.52, z-score = -1.47, R) >droEre2.scaffold_4784 21019180 99 - 25762168 -AAGGGAUCAACCUA-UUAAACUCCAGUUCCUAUGUUUGCUUUUCCGGUCCGGCCCCUUUGGAGAUGCGCAAACAUCUUCGCCCGGGGCUGGACAAUCUAU----- -..((((.(((.(..-...(((....))).....).)))...)))).((((((((((..((((((((......))))))))...)))))))))).......----- ( -34.90, z-score = -2.68, R) >droAna3.scaffold_13337 10179478 80 - 23293914 AACGGAAUAAA--UAUUU---CCCACAGUACUUUGUUGG-UUUUCCGGUCCGGCCCCUGAGAUGA-ACGGUCACAUGCUGGACCCAU------------------- ...((((....--.....---((.((((....)))).))-..))))((((((((...((.(((..-...))).)).))))))))...------------------- ( -22.30, z-score = -0.94, R) >droWil1.scaffold_180708 8943450 102 + 12563649 -AAGAAAUUGAUUUACAU---UUUGGUGAACGACACGAGAAUCGCCGUAUCAGUUUCCCGUGACGCCCUACCGCACCGCCAACCAGCUCAUUUCUCUGUCAUCUAU -..(((((((((....((---(((.(((.....))).)))))......)))))))))..(((((((......))...((......))..........))))).... ( -19.80, z-score = -0.89, R) >consensus AAGGAAAU_AACCUAUUUAAACUCAAGUUCCUAUGUUUGCUUUUCCGGUCCGGCCCCUUUGGAGAUUCGAACACAUCUUCGCCCGGGGUUGGACAAUCUAU_____ ...............................................((((((((((...........................))))))))))............ ( -8.68 = -10.64 + 1.96)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:02 2011