
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,332,785 – 21,332,893 |
| Length | 108 |
| Max. P | 0.651761 |

| Location | 21,332,785 – 21,332,893 |
|---|---|
| Length | 108 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.50 |
| Shannon entropy | 0.59072 |
| G+C content | 0.47934 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -17.89 |
| Energy contribution | -17.48 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
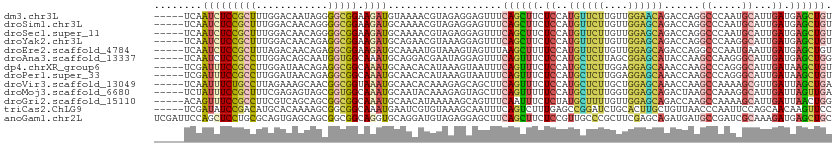
>dm3.chr3L 21332785 108 + 24543557 -----UCAAUCUCCGCUUUGGACAAUAGGGGCGGAAGAUGUAAAACGUAGAGGAGUUUCAGCUUCUCCAUGUUCUUGUUGGAACAGACCAGGCCCAAUGCAUUGAUGAGCUGU -----(((.((((((((((........)))))))).(((((..(((((.(((((((....))))))).)))))...(((((..(......)..)))))))))))))))..... ( -31.20, z-score = -0.55, R) >droSim1.chr3L 20679764 108 + 22553184 -----UCAAUCUCCGCUUUGGACAACAGGGGCGGAAGAUGCAAAACGUAGAGGAGUUUCAGCUUCUCCAUGUUCUUGUUGGAGCAGACCAGGCCCAAUGCAUUGAUGAGCUGU -----(((.((((((((((........)))))))).(((((..(((((.(((((((....))))))).)))))...(((((.((.......)))))))))))))))))..... ( -34.90, z-score = -0.86, R) >droSec1.super_11 1270072 108 + 2888827 -----UCAAUCUCCGCUUUGGACAACAGGGGCGGAAGAUGCAAAACGUAGAGGAGUUUCAGCUUCUCCAUGUUCUUGUUGGAGCAGACCAGGCCCAAUGCAUUGAUGAGCUGU -----(((.((((((((((........)))))))).(((((..(((((.(((((((....))))))).)))))...(((((.((.......)))))))))))))))))..... ( -34.90, z-score = -0.86, R) >droYak2.chr3L 5002913 108 - 24197627 -----UCAAUCUCCGCUUUGGACAACAGAGGCGGAAGAUGCAGAACGUAAAGGAGUUUCAGCUUCUCCAUGUUCUUGUUGGAGCAGACCAGGCCCAAGGCAUUGAUGAGCUGU -----(((.((((((((((........)))))))).((((((((((((..((((((....))))))..)))))))..((((.((.......)))))).))))))))))..... ( -35.40, z-score = -1.16, R) >droEre2.scaffold_4784 21001458 108 + 25762168 -----UCAAUCUCCGCUUUAGACAACAGAGGCGGAAGAUGCAAAAUGUAAAGUAGUUUAAGCUUUUCCAUGUUCUUGUUGGAGCAGACCAGGCCCAAUGAAUUGAUGAGCUGU -----...(((((((((((........))))))).))))....................(((((.((...((((..(((((.((.......))))))))))).)).))))).. ( -27.60, z-score = -0.82, R) >droAna3.scaffold_13337 10160125 108 + 23293914 -----UCAAUCUCCGCCUUGGACAGCAAUGGUGGCAAAUGCAGGACGAAUAGGAGUUUCAGUUUCUCCAUGCUCUUAGCGGAGCAUACCAAGCCAAGGGCAUUGAUGAGCUGG -----.....((((...(((..(.(((.((....))..))).)..)))...))))...((((((.((.((((((((.((((......))..)).)))))))).)).)))))). ( -31.90, z-score = -0.04, R) >dp4.chrXR_group6 12685395 108 + 13314419 -----UCGAUUUCCGCCUUGGAUAACAGAGGCGGCAAAUGCAACACAUAAAGUAAUUUCAGUUUCUCCAUGCUCUUGGAGGAGCAAACCAAGCCCAGGGCAUUGAUAAGCUGU -----...(((((((((((........))))))).))))...................((((((.((.(((((((.((.((......))...)).))))))).)).)))))). ( -33.50, z-score = -2.07, R) >droPer1.super_33 706940 108 + 967471 -----UCGAUUUCCGCCUUGGAUAACAGAGGCGGCAAAUGCAACACAUAAAGUAAUUUCAGUUUCUCCAUGCUCUUGGAGGAGCAAACCAAGCCCAGGGCAUUGAUAAGCUGU -----...(((((((((((........))))))).))))...................((((((.((.(((((((.((.((......))...)).))))))).)).)))))). ( -33.50, z-score = -2.07, R) >droVir3.scaffold_13049 9518781 108 + 25233164 -----UCAAUUUCUGCCUUAGAAAGCAACGGCGGUAAAUGCAACACAAAGAGCAGCUUCAGUUUCUCCAUGCUCUUGCUGGAGCAAACCAAGCCAAAAGCGUUGAUUAGCUGA -----...(((((((((............))))).)))).............(((((((((((((((((.((....))))))).))))...((.....))..)))..))))). ( -26.80, z-score = 0.22, R) >droMoj3.scaffold_6680 13519997 108 - 24764193 -----UCUAUUUCCGCUUUCGAGAGUAGCGGUGGCAAAUGCAAUACAAAGAGUAGCUUCAGUUUUUCCAUGCUCUUGGUGGAGCAGACUAAGCCAAAGGCAUUGAUUAGUUGA -----.(((((((((((..(....).)))))..((....))........))))))..(((..(..((.((((..(((((..((....))..)))))..)))).))..)..))) ( -30.20, z-score = -0.83, R) >droGri2.scaffold_15110 11498041 108 - 24565398 -----ACAGUUUCCGCCUUCGUCAGCAGCGGCGGCAAAUGCAACAUAAAAAGCAGUUUCAAUUUCUCUAUGCUUUUGUUGGAGCAGACCAAGCCAAAAGCAUUGAUUAACUGG -----.(((((...(((.((((.....)))).)))...(((..........)))...........((.(((((((((((((......))))..))))))))).))..))))). ( -27.30, z-score = -0.68, R) >triCas2.ChLG9 2582678 108 + 15222296 -----UCGAUAUCCGACAUGCACAAAAGCGGCGGCAAAUGAAUCGUGUAAAGCAAUUUCAGUCUUUGAGCCGGAUCUGCACUUGCUGUUAACCCAAUUCCAGCAACAAGUUCC -----(((.....)))...((......))(((..((((((((...((.....))..))))...)))).)))(((.((....((((((..(......)..))))))..)).))) ( -21.40, z-score = 0.60, R) >anoGam1.chr2L 22470554 113 - 48795086 UCGAUUCCAGCUCCUGCGCAGUGAGCAGCGGCGGCAGGUGCAGGAUGUAGAGGAGCUUCAGCUUCUCCGUUGCCCGCUUCGAGCAGAUGAUGCCGAUCGCAAAGAUGAGCUGC .......((((((.(((.......)))(((((((((((.(((..(((..(((((((....)))))))))))))))((.....))......))))).))))......)))))). ( -43.10, z-score = 0.17, R) >consensus _____UCAAUCUCCGCCUUGGACAACAGCGGCGGCAAAUGCAAAACGUAAAGGAGUUUCAGUUUCUCCAUGCUCUUGUUGGAGCAGACCAAGCCCAAGGCAUUGAUGAGCUGU ........(((((((((............))))).))))...................((((((.((..((((((....))))))......((.....))...)).)))))). (-17.89 = -17.48 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:02 2011