| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,313,356 – 21,313,461 |
| Length | 105 |
| Max. P | 0.584226 |

| Location | 21,313,356 – 21,313,461 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.13 |
| Shannon entropy | 0.49725 |
| G+C content | 0.57404 |
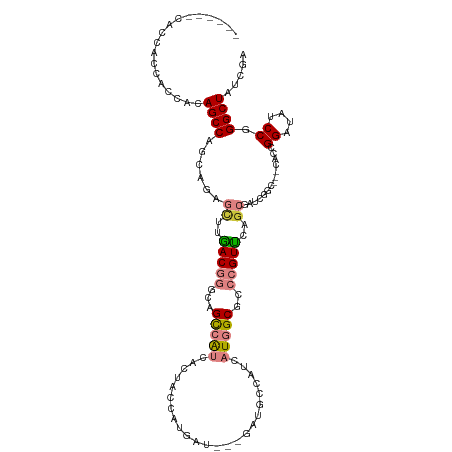
| Mean single sequence MFE | -33.71 |
| Consensus MFE | -14.59 |
| Energy contribution | -15.25 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
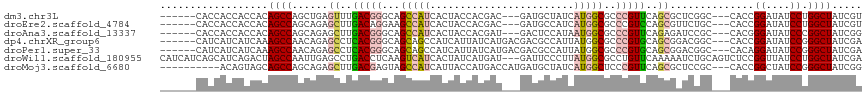
>dm3.chr3L 21313356 105 + 24543557 ------CACCACCACCACAGCCAGCUGAGUUUGACGGGCAGCCAUCACUACCACGAC---GAUGCUAUCAUGGCGCCCGUUCAGCGCUCGGC---CACCGGAUAUCCUGGCUAUCGU ------............(((((((((((((((((((((.(((((...((.((....---..)).))..)))))))))).)))).)))))))---....((....)))))))..... ( -37.80, z-score = -2.38, R) >droEre2.scaffold_4784 20981691 105 + 25762168 ------CACCACCACCACAGCCAGCAGAGCUUGACAGGAAGCCAUCACUACCACGAC---GAUGCCAUCAUGGCGCCCGUCCAGCGUUCUGC---CACCGGAUAUCCUGGCUAUCGU ------............(((((((((((((((...((.((......)).))..(((---(.((((.....))))..))))))).)))))))---....((....)))))))..... ( -32.20, z-score = -2.05, R) >droAna3.scaffold_13337 10142226 105 + 23293914 ------CACCACCACCACAGCCAGCAGAGCUUGACGGGCAGCCAUCACUACCACGAU---GACUCCAUAAUGGCGCCCGUUCAGAGAUCCGC---CACGGGAUAUCCCGGCUAUCGG ------........((..((((.((.((.((.(((((((.(((((..........((---(....))).)))))))))))).))...)).))---...(((....)))))))...)) ( -35.30, z-score = -2.80, R) >dp4.chrXR_group6 12667135 108 + 13314419 ------CAUCAUCAUCAAAGCCAACAGAGCCUCACGGGCAGCAGCCAUCAUUAUCAUGACGACGCCAUUAUGGCGCCCGUGCAGCGGACGGC---CACCGGAUAUCCGGGCUAUCGA ------.............(((..(...((..(((((((....(.(.((((....)))).).)(((.....))))))))))..)).)..)))---..((((....))))........ ( -35.70, z-score = -1.32, R) >droPer1.super_33 688644 108 + 967471 ------CAUCAUCAUCAAAGCCAACAGAGCCUCACGGGCAGCAGCCAUCAUUAUCAUGACGACGCCAUUAUGGCGCCCGUGCAGCGGACGGC---CACAGGAUAUCCGGGCUAUCGA ------......................((..(((((((....(.(.((((....)))).).)(((.....))))))))))..)).((..((---(...((....)).)))..)).. ( -34.70, z-score = -1.35, R) >droWil1.scaffold_180955 278309 114 + 2875958 CAUCAUCAGCAUCAGACUAGCCAAUUGAGCCUGACCUCAAGUCAUCACUAUCAUGAU---GAUUCCCUUAUGGCGCCUGUUCAAAAAUCUGCAGUCUCCGGUUAUCCUGGCUAUCGA ..............((.((((((....((((........((((((((......))))---)))).......((.(.((((..........)))).).))))))....)))))))).. ( -26.80, z-score = -0.90, R) >droMoj3.scaffold_6680 13502190 104 - 24764193 ----------ACAGUAGCAGCCAGCAGAGCUUGACGAGUAGCCAUCAUUACCAUGACCAUGAUGCUAUCAUGGCUCCCGUUCAGCGCUCCGC---CACCGGCUAUCCGGGCUAUCGG ----------.(.(((((.....((.((((((((((.(.((((.((((....))))..(((((...))))))))).))).)))).)))).))---..((((....))))))))).). ( -33.50, z-score = -1.03, R) >consensus ______CACCACCACCACAGCCAGCAGAGCUUGACGGGCAGCCAUCACUACCAUGAU___GAUGCCAUCAUGGCGCCCGUUCAGCGAUCGGC___CACCGGAUAUCCGGGCUAUCGA ..................((((......((..(((((...(((((........................)))))..)))))..))..............((....)).))))..... (-14.59 = -15.25 + 0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:42:00 2011