
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,235,524 – 21,235,621 |
| Length | 97 |
| Max. P | 0.845623 |

| Location | 21,235,524 – 21,235,621 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Shannon entropy | 0.43756 |
| G+C content | 0.51017 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -13.85 |
| Energy contribution | -13.55 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
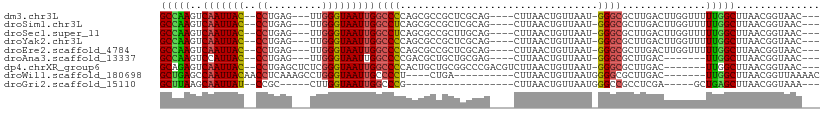
>dm3.chr3L 21235524 97 - 24543557 GCCAAGUCAAUUAC--CCUGAG---UUGGGUAAUUGGCCCCAGCGCCGCUCGCAG----CUUAACUGUUAAU-GGGCGCUUGACUUGGUUUUUGGCUUAACGGUAAC--- ((((((((((((((--((....---..))))))))))).(.((((((.(..((((----.....))))....-))))))).).........)))))...........--- ( -37.10, z-score = -2.52, R) >droSim1.chr3L 20595464 97 - 22553184 GCCAAGUCAAUUAC--CCUGAG---UUGGGUAAUUGGCCUCAGCGCCGCUCGCAG----CUUAACUGUUAAU-GGGCGCUUGACUUGGUUUUUGGCUUAACGGUAAC--- ((((((((((((((--((....---..))))))))))).((((((((.(..((((----.....))))....-)))))).)))........)))))...........--- ( -37.60, z-score = -3.03, R) >droSec1.super_11 1188960 97 - 2888827 GCCAAGUCAAUUAC--CCUGAG---UUGGGUAAUUGGCCUCAGCGCCGCUUGCAG----CUUAACUGUUAAU-GGGCGCUUGACUUGGUUUUUGGCUUAACGGUAAC--- ((((((((((((((--((....---..))))))))))).((((((((.(..((((----.....))))....-)))))).)))........)))))...........--- ( -38.30, z-score = -3.26, R) >droYak2.chr3L 4921012 97 + 24197627 GCCAAGUCAAUUAC--CCUGAG---UUGGGUAAUUGGCCCCAGCGCCGCUCGCAG----CUUAACUGUUAAU-GGGCGCUUGACUUGGUUUUUGGCUUAACGGUAAC--- ((((((((((((((--((....---..))))))))))).(.((((((.(..((((----.....))))....-))))))).).........)))))...........--- ( -37.10, z-score = -2.52, R) >droEre2.scaffold_4784 20919531 97 - 25762168 GCCAAGUCAAUUAC--CCUGAG---UUGGGUAAUUGGCCCCAGCGCCGCUCGCAG----CUUAACUGUUAAU-GGGCGCUUGACUUGGUUUUUGGCUUAACGGUAAC--- ((((((((((((((--((....---..))))))))))).(.((((((.(..((((----.....))))....-))))))).).........)))))...........--- ( -37.10, z-score = -2.52, R) >droAna3.scaffold_13337 14839674 90 + 23293914 GCCAAGUCCAUUAC--CCUGAG---UUGGGUAAUUGGCCCCGACGCUGCUGCGAG----CUUAACUGUUAAU-GGGCGCUUGAC-------UUGGCUUAACGGUAAC--- ((((((((.(((((--((....---..))))))).(((((((.(((....)))((----(......)))..)-))).))).)))-------)))))...........--- ( -35.30, z-score = -3.19, R) >dp4.chrXR_group6 12606500 97 - 13314419 GCAGAGUCAAUUAC--CCUGAGCUCUCGGGUAAUUGGCCCCACUGCUGCGGCCCGACGUCUUAACUGUUAAU-GGGCGCUUGAC-------UUGGCUUAACGGUAAC--- ((((.(((((((((--((.((....)))))))))))))....))))..(((((((....(......)....)-))))(((....-------..)))....)).....--- ( -32.10, z-score = -1.30, R) >droWil1.scaffold_180698 5661594 89 + 11422946 GCUGAGCCAAUUACAACCUCAAAGCCUGGGUAAUUGCCCCU----CUGA----------CUUAACUGUUAAUGGGGCGCUUGAC-------UUGGCUUAACGGUUAAAAC ..............((((...(((((.((.(((.((((((.----.(((----------(......))))..)))))).))).)-------).)))))...))))..... ( -28.80, z-score = -3.06, R) >droGri2.scaffold_15110 12545724 77 + 24565398 GCUUAAGCAAUUAU--CCGC-----CUUGGUAAUUGGCCCG------------------CUUAACUGUUAAUGGGCCGCCUCGA-----GCUGAGCUUAACGGUAAA--- (((((.((((((((--(...-----...)))))))((((((------------------.(((.....))))))))).......-----)))))))...........--- ( -21.90, z-score = -0.51, R) >consensus GCCAAGUCAAUUAC__CCUGAG___UUGGGUAAUUGGCCCCAGCGCCGCUCGCAG____CUUAACUGUUAAU_GGGCGCUUGACUUGGUUUUUGGCUUAACGGUAAC___ (((((..(((((((..((.........)))))))))((((.................................))))..............))))).............. (-13.85 = -13.55 + -0.29)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:55 2011