
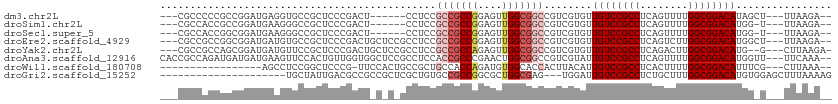
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,996,402 – 5,996,503 |
| Length | 101 |
| Max. P | 0.999823 |

| Location | 5,996,402 – 5,996,500 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 67.47 |
| Shannon entropy | 0.64384 |
| G+C content | 0.64417 |
| Mean single sequence MFE | -42.44 |
| Consensus MFE | -24.77 |
| Energy contribution | -25.49 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.998867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
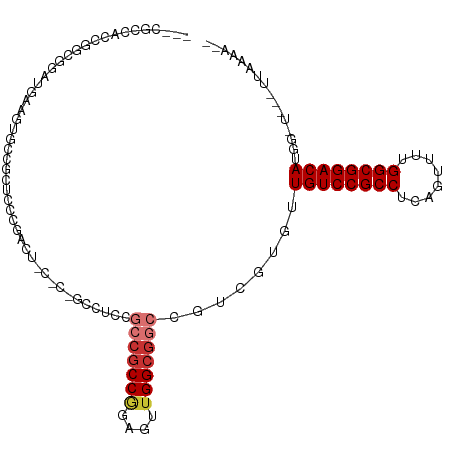
>dm3.chr2L 5996402 98 + 23011544 ---CGCCCCCGCCGGAUGAGGUGCCGCUCCCGACU------CCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUUUUGGCGGACAUAGCU---UUAAGA-- ---.......((...((((.(.(((((...(((((------((.........))))))))))))).))))..((((((((........))))))))..)).---......-- ( -40.40, z-score = -1.19, R) >droSim1.chr2L 5784762 97 + 22036055 ---CGCCACCGCCGGAUGAAGGGCCGCUCCCGACU------CCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUUUUGGCGGACAUGG-U---UUAAGA-- ---.((((.......((((..((((((...(((((------((.........))))))))))))).))))..((((((((........)))))))))))-)---......-- ( -43.80, z-score = -1.48, R) >droSec1.super_5 4067447 97 + 5866729 ---CGCCACCGGCGGAUGAAGGGCCGCUCCCGACU------CCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUUUUGGCGGACAUGG-U---UUAAGA-- ---((((((((((((.((.((((.((....)).).------))).)))))))))...)))))((((......((((((((........)))))))))))-)---......-- ( -45.20, z-score = -1.54, R) >droEre2.scaffold_4929 6087105 104 + 26641161 ---CGCCGCCGGCGGAUGAUGUGCCGCUCCCGACUGCUCCGCCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUCUUGGCGGACAUGGCU---UUAAGA-- ---((((.(((((((..((.(((..((........))..))).))..)))))))....))))((((......((((((((........)))))))))))).---......-- ( -48.50, z-score = -1.49, R) >droYak2.chr2L 9126614 103 - 22324452 ---CGCCGCCAGCGGAUGAUGUUCCGCUCCCGACUGCUCCGCCUCCGCCGCCAGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGACUUGGCGGACAUG--G---CUUAAGA- ---....(((((((((......)))))............(((..(.((((((......)))))).)..))).((((((((........))))))))))--)---)......- ( -47.00, z-score = -2.38, R) >droAna3.scaffold_12916 11411352 107 + 16180835 CACCGCCAGAUGAUGAUGAAGUUCCACUGUUGGUGGCUCCGCCUCCACCGCCCGAACUGGCGGCCGUCGUAUUGUCCGCCUCAGUUUUGGCGGACAUGGUU---UUCAAA-- ....((((....((((((............(((.(((...))).)))(((((......))))).))))))..((((((((........)))))))))))).---......-- ( -40.60, z-score = -2.10, R) >droWil1.scaffold_180708 4598033 89 - 12563649 -----------------AGCCUCCGGCUCCCG-UUCCACUGCCGCUGCCACCAGAUGUGGCACCACUUACAUUGUCCGCCUCACUUUUGGCGGACAUUUCG---CUUAAA-- -----------------(((...((((.....-.......)))).((((((.....))))))..........((((((((........))))))))....)---))....-- ( -30.50, z-score = -3.78, R) >droGri2.scaffold_15252 2871953 88 + 17193109 ---------------------UGCUAUUGACGCCGCCGCUCGCUGUGCCGCCGGCGCUGGCGAG---UGGAUUGUCCGCCUCUGCUUUGGCGGACAUGUGGAGCUUUAAAAG ---------------------.(((.....(((..((((((((((((((...))))).))))))---)))..((((((((........)))))))).))).)))........ ( -43.50, z-score = -3.58, R) >consensus ___CGCCACCGGCGGAUGAAGUGCCGCUCCCGACU_C_C_GCCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUUUUGGCGGACAUGG_U___UUAAAA__ ..............................................(((((((....)))))))........((((((((........))))))))................ (-24.77 = -25.49 + 0.72)
| Location | 5,996,402 – 5,996,500 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 67.47 |
| Shannon entropy | 0.64384 |
| G+C content | 0.64417 |
| Mean single sequence MFE | -45.58 |
| Consensus MFE | -27.14 |
| Energy contribution | -27.24 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5996402 98 - 23011544 --UCUUAA---AGCUAUGUCCGCCAAAACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGG------AGUCGGGAGCGGCACCUCAUCCGGCGGGGGCG--- --......---.(((.((((((((........))))))))...((.((((((((......)))))).((((------.((((....)))).))))...)).))..))).--- ( -48.60, z-score = -3.08, R) >droSim1.chr2L 5784762 97 - 22036055 --UCUUAA---A-CCAUGUCCGCCAAAACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGG------AGUCGGGAGCGGCCCUUCAUCCGGCGGUGGCG--- --......---.-(((((((((((........)))))))....((.((((((((......))))))(((((------.((((....)))))))))...)).))))))..--- ( -48.50, z-score = -3.02, R) >droSec1.super_5 4067447 97 - 5866729 --UCUUAA---A-CCAUGUCCGCCAAAACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGG------AGUCGGGAGCGGCCCUUCAUCCGCCGGUGGCG--- --......---.-...((((((((........))))))))..........(((((...(((((((.(((((------.((((....)))))))))..))))))))))))--- ( -50.70, z-score = -3.77, R) >droEre2.scaffold_4929 6087105 104 - 26641161 --UCUUAA---AGCCAUGUCCGCCAAGACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGGCGGAGCAGUCGGGAGCGGCACAUCAUCCGCCGGCGGCG--- --......---.(((.((((((((........))))))))......(.((((((......)))))).).((((((...((((....))))......)))))))))....--- ( -53.00, z-score = -3.35, R) >droYak2.chr2L 9126614 103 + 22324452 -UCUUAAG---C--CAUGUCCGCCAAGUCUGAGGCGGACAACACGACGGCCGCCAACUCUGGCGGCGGAGGCGGAGCAGUCGGGAGCGGAACAUCAUCCGCUGGCGGCG--- -......(---(--(.((((((((........))))))))......(.(((((((....))))))).).)))......((((..((((((......))))))..)))).--- ( -53.00, z-score = -3.58, R) >droAna3.scaffold_12916 11411352 107 - 16180835 --UUUGAA---AACCAUGUCCGCCAAAACUGAGGCGGACAAUACGACGGCCGCCAGUUCGGGCGGUGGAGGCGGAGCCACCAACAGUGGAACUUCAUCAUCAUCUGGCGGUG --......---.....((((((((........))))))))........((((((((....(..(((((((......((((.....))))..)))))))..)..)))))))). ( -45.50, z-score = -3.22, R) >droWil1.scaffold_180708 4598033 89 + 12563649 --UUUAAG---CGAAAUGUCCGCCAAAAGUGAGGCGGACAAUGUAAGUGGUGCCACAUCUGGUGGCAGCGGCAGUGGAA-CGGGAGCCGGAGGCU----------------- --.....(---(....((((((((........)))))))).......((.((((((.....)))))).)))).......-....((((...))))----------------- ( -32.80, z-score = -1.90, R) >droGri2.scaffold_15252 2871953 88 - 17193109 CUUUUAAAGCUCCACAUGUCCGCCAAAGCAGAGGCGGACAAUCCA---CUCGCCAGCGCCGGCGGCACAGCGAGCGGCGGCGUCAAUAGCA--------------------- ........((.((...((((((((........))))))))..((.---(((((..(.(((...))).).))))).)).)).))........--------------------- ( -32.50, z-score = -1.32, R) >consensus __UCUUAA___A_CCAUGUCCGCCAAAACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGGC_G_G_AGUCGGGAGCGGCACUUCAUCCGCCGGUGGCG___ ................((((((((........))))))))......(.((((((......)))))).)............................................ (-27.14 = -27.24 + 0.10)
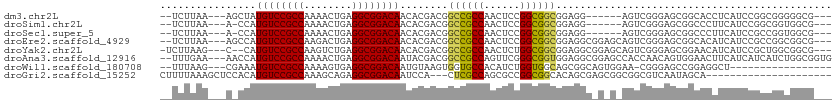
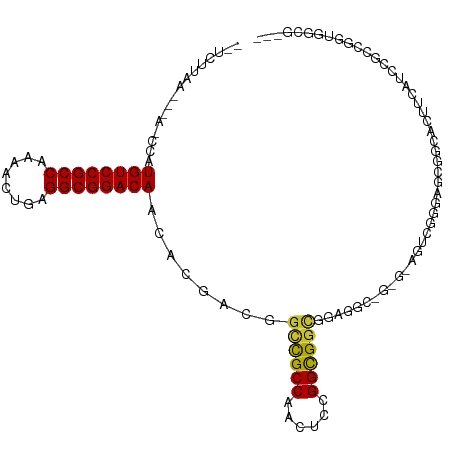
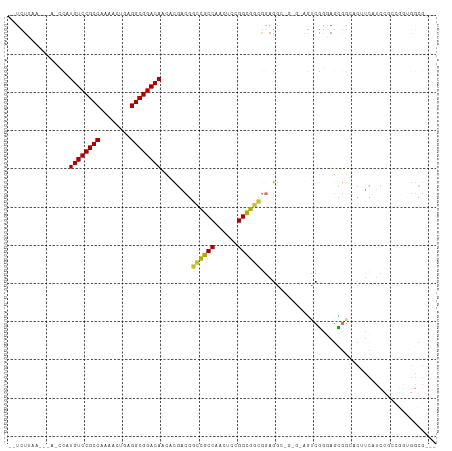
| Location | 5,996,403 – 5,996,503 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.62 |
| Shannon entropy | 0.52237 |
| G+C content | 0.63411 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -25.99 |
| Energy contribution | -27.03 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.991994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
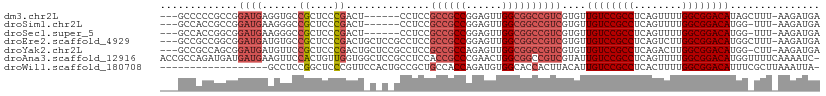
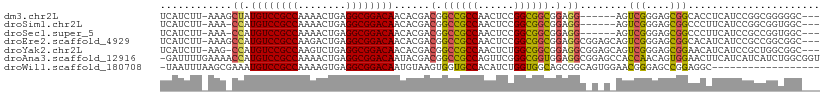
>dm3.chr2L 5996403 100 + 23011544 ---GCCCCCGCCGGAUGAGGUGCCGCUCCCGACU------CCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUUUUGGCGGACAUAGCUUU-AAGAUGA ---......((...((((.(.(((((...(((((------((.........))))))))))))).))))..((((((((........))))))))..))...-....... ( -40.40, z-score = -1.20, R) >droSim1.chr2L 5784763 99 + 22036055 ---GCCACCGCCGGAUGAAGGGCCGCUCCCGACU------CCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUUUUGGCGGACAUGG-UUU-AAGAUGA ---((((.......((((..((((((...(((((------((.........))))))))))))).))))..((((((((........)))))))))))-)..-....... ( -43.50, z-score = -1.44, R) >droSec1.super_5 4067448 99 + 5866729 ---GCCACCGGCGGAUGAAGGGCCGCUCCCGACU------CCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUUUUGGCGGACAUGG-UUU-AAGAUGA ---(((...((((((.((.(((.....)))...)------).))))))((((....)))))))((((....((((((((........))))))))...-...-..)))). ( -45.20, z-score = -1.57, R) >droEre2.scaffold_4929 6087106 106 + 26641161 ---GCCGCCGGCGGAUGAUGUGCCGCUCCCGACUGCUCCGCCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUCUUGGCGGACAUGGCUUU-AAGAUGA ---(((((((((((........))))).......((((((.(......).)))))).))))))((((..((((((((((........))))))))..))...-..)))). ( -47.50, z-score = -1.34, R) >droYak2.chr2L 9126615 105 - 22324452 ---GCCGCCAGCGGAUGAUGUUCCGCUCCCGACUGCUCCGCCUCCGCCGCCAGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGACUUGGCGGACAUGG-CUU-AAGAUGA ---...(((((((((......)))))............(((..(.((((((......)))))).)..))).((((((((........)))))))))))-)..-....... ( -47.00, z-score = -2.43, R) >droAna3.scaffold_12916 11411353 109 + 16180835 ACCGCCAGAUGAUGAUGAAGUUCCACUGUUGGUGGCUCCGCCUCCACCGCCCGAACUGGCGGCCGUCGUAUUGUCCGCCUCAGUUUUGGCGGACAUGGUUUUCAAAAUC- ...((((....((((((............(((.(((...))).)))(((((......))))).))))))..((((((((........))))))))))))..........- ( -40.60, z-score = -2.22, R) >droWil1.scaffold_180708 4598034 91 - 12563649 ------------------GCCUCCGGCUCCCGUUCCACUGCCGCUGCCACCAGAUGUGGCACCACUUACAUUGUCCGCCUCACUUUUGGCGGACAUUUCGCUUAAAUUA- ------------------......(((............)))((((((((.....))))))..........((((((((........))))))))....))........- ( -30.00, z-score = -4.05, R) >consensus ___GCCACCGGCGGAUGAAGUGCCGCUCCCGACU_C_C_GCCUCCGCCGCCGGAGUUGGCGGCCGUCGUGUUGUCCGCCUCAGUUUUGGCGGACAUGG_UUU_AAGAUGA .............((((......((....))..............((((((......))))))))))....((((((((........))))))))............... (-25.99 = -27.03 + 1.04)
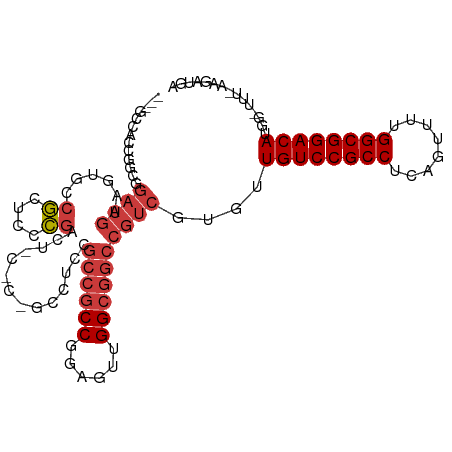
| Location | 5,996,403 – 5,996,503 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.62 |
| Shannon entropy | 0.52237 |
| G+C content | 0.63411 |
| Mean single sequence MFE | -46.37 |
| Consensus MFE | -29.77 |
| Energy contribution | -29.39 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
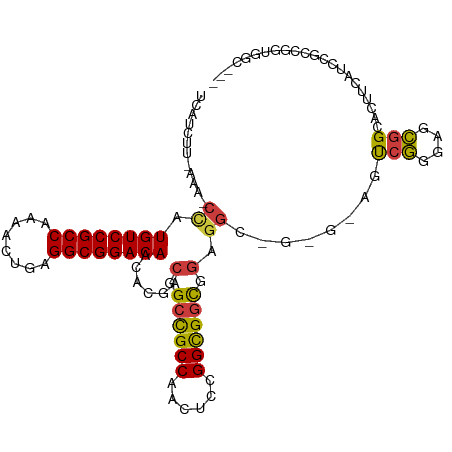
>dm3.chr2L 5996403 100 - 23011544 UCAUCUU-AAAGCUAUGUCCGCCAAAACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGG------AGUCGGGAGCGGCACCUCAUCCGGCGGGGGC--- .......-...(((.((((((((........))))))))...((.((((((((......)))))).((((------.((((....)))).))))...)).))..)))--- ( -46.90, z-score = -2.79, R) >droSim1.chr2L 5784763 99 - 22036055 UCAUCUU-AAA-CCAUGUCCGCCAAAACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGG------AGUCGGGAGCGGCCCUUCAUCCGGCGGUGGC--- .......-...-(((((((((((........)))))))....((.((((((((......))))))(((((------.((((....)))))))))...)).)))))).--- ( -48.50, z-score = -3.11, R) >droSec1.super_5 4067448 99 - 5866729 UCAUCUU-AAA-CCAUGUCCGCCAAAACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGG------AGUCGGGAGCGGCCCUUCAUCCGCCGGUGGC--- .......-...-(((((((((((........)))))))....((.((((((((......))))))(((((------.((((....)))))))))...)).)))))).--- ( -48.50, z-score = -3.24, R) >droEre2.scaffold_4929 6087106 106 - 26641161 UCAUCUU-AAAGCCAUGUCCGCCAAGACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGGCGGAGCAGUCGGGAGCGGCACAUCAUCCGCCGGCGGC--- .......-...(((.((((((((........))))))))......(.((((((......)))))).).((((((...((((....))))......)))))))))...--- ( -53.00, z-score = -3.59, R) >droYak2.chr2L 9126615 105 + 22324452 UCAUCUU-AAG-CCAUGUCCGCCAAGUCUGAGGCGGACAACACGACGGCCGCCAACUCUGGCGGCGGAGGCGGAGCAGUCGGGAGCGGAACAUCAUCCGCUGGCGGC--- .......-..(-((.((((((((........))))))))......(.(((((((....))))))).).)))......((((..((((((......))))))..))))--- ( -51.30, z-score = -3.21, R) >droAna3.scaffold_12916 11411353 109 - 16180835 -GAUUUUGAAAACCAUGUCCGCCAAAACUGAGGCGGACAAUACGACGGCCGCCAGUUCGGGCGGUGGAGGCGGAGCCACCAACAGUGGAACUUCAUCAUCAUCUGGCGGU -..............((((((((........))))))))........((((((((....(..(((((((......((((.....))))..)))))))..)..)))))))) ( -44.80, z-score = -2.99, R) >droWil1.scaffold_180708 4598034 91 + 12563649 -UAAUUUAAGCGAAAUGUCCGCCAAAAGUGAGGCGGACAAUGUAAGUGGUGCCACAUCUGGUGGCAGCGGCAGUGGAACGGGAGCCGGAGGC------------------ -........((....((((((((........)))))))).......((.((((((.....)))))).)))).......(((...))).....------------------ ( -31.60, z-score = -2.01, R) >consensus UCAUCUU_AAA_CCAUGUCCGCCAAAACUGAGGCGGACAACACGACGGCCGCCAACUCCGGCGGCGGAGGC_G_G_AGUCGGGAGCGGCACUUCAUCCGCCGGUGGC___ ...............((((((((........))))))))......(.((((((......)))))).)...........(((....)))...................... (-29.77 = -29.39 + -0.38)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:15 2011