| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,231,394 – 21,231,493 |
| Length | 99 |
| Max. P | 0.646225 |

| Location | 21,231,394 – 21,231,493 |
|---|---|
| Length | 99 |
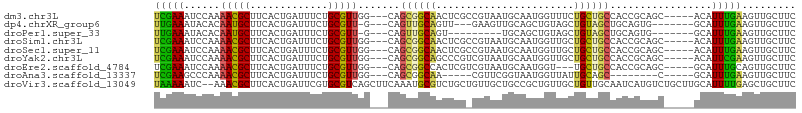
| Sequences | 9 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.54 |
| Shannon entropy | 0.51993 |
| G+C content | 0.49531 |
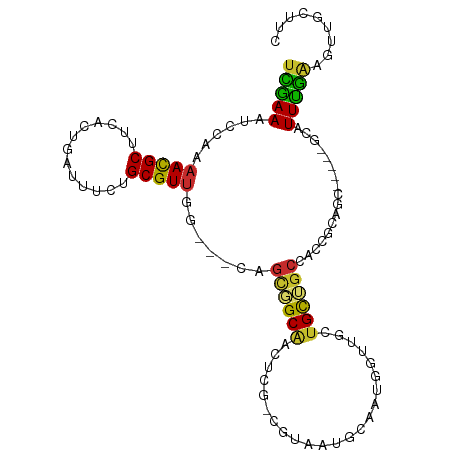
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -9.96 |
| Energy contribution | -9.23 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21231394 99 - 24543557 UCGAAAUCCAAAACGCUUCACUGAUUUCUGCGUUGG---CAGCGGCAACUCGCCGUAAUGCAAUGGUUUCUGCUGCCACCGCAGC-----ACAUUUGAAGUUGCUUC ..............((((((.......(((((.(((---((((((.((((.((......))...)))).))))))))).))))).-----.....))))))...... ( -34.22, z-score = -2.66, R) >dp4.chrXR_group6 12601066 93 - 13314419 UUGAAAUACACAAUGCUUCACUGAUUUCUGCGUU-G---CAGUUGCAGUU---GAAGUUGCAGCUGUAGCUGUAGCUGCAGUG-------GCAUUUGAAGUUGCUUC ..............((((((.((....(((((((-(---(((((((((((---(......)))))))))))))))).))))..-------.))..))))))...... ( -36.20, z-score = -2.59, R) >droPer1.super_33 622205 87 - 967471 UUGAAAUACACAAUGCUUCACUGAUUUCUGCGUU-G---CAGUUGCAGU---------UGCAGCUGUAGCUGUAGCUGCAGUG-------GCAUUUGAAGUUGCUUC ..............((((((.((....(((((((-(---((((((((((---------....)))))))))))))).))))..-------.))..))))))...... ( -32.60, z-score = -2.00, R) >droSim1.chr3L 20591340 99 - 22553184 UCGAAAUCCAAAACGCUUCACUGAUUUCUGCGUUGG---CAGCGGCAACUCGCCGUAAUGCAAUGGUUGCUGCUGCCACCGCAGC-----ACAUUUGAAGUUGCUUC ..............((((((.......(((((.(((---(((((((((((.((......))...)))))))))))))).))))).-----.....))))))...... ( -40.82, z-score = -4.17, R) >droSec1.super_11 1184837 99 - 2888827 UCGAAAUCCAAAACGCUUCACUGAUUUCUGCGUUGG---CAGCGGCAACUCGCCGUAAUGCAAUGGUUGCUGCUGCCACCGCAGC-----ACAUUUGAAGUUGCUUC ..............((((((.......(((((.(((---(((((((((((.((......))...)))))))))))))).))))).-----.....))))))...... ( -40.82, z-score = -4.17, R) >droYak2.chr3L 4903919 99 + 24197627 UCGAAAUCCAAAACGCUUCACUGAUUUCUGCGUUGG---CAGCGGCAGCCCGUCGUAAUGCAAUGGUUGCUGCUGCCACCGCAGC-----ACAUUCGAAGUUGCUUC ..............(((((........(((((.(((---(((((((((((.((.(.....).)))))))))))))))).))))).-----......)))))...... ( -39.56, z-score = -3.37, R) >droEre2.scaffold_4784 20915405 96 - 25762168 UCGAAAUCCAAAACGCUUCACUGAUUUCUGCGUUGG---CAGCGGCCACUCGUCGUAAUGCAAUGGU---UGCUGCCACCGCAGC-----GCAUUUGCAGUUGCUUC ..............((...((((....(((((.(((---(((((((((..(((....)))...))))---)))))))).))))).-----.......)))).))... ( -34.50, z-score = -2.17, R) >droAna3.scaffold_13337 14834973 86 + 23293914 UCGAAGCCCAAAACGCUUCACUGAUUUCUGCGUUGG---CAGCGGCAA-----CGUUCGGUAAUGGUUAUUGCAGC--------C-----GCAUUUGAAGUUGCUUC ..(((((.......)))))........((((....)---))).(((((-----(.(((((...(((((.....)))--------)-----)...))))))))))).. ( -23.80, z-score = -0.01, R) >droVir3.scaffold_13049 20472529 105 - 25233164 UAAAAAUC--AAACGCUUCACUGAUUCGUGCGUCAGCUUCAAAUGCGUCUGCUGUUGCUGCCGCUGUUGCUGUUGCAAUCAUGUCUGCUUGCAUUUUGAGCUGCUUC ........--..((((.............))))((((((.(((((((...((.(((((.((.((....)).)).))))).......)).))))))).)))))).... ( -27.42, z-score = -0.64, R) >consensus UCGAAAUCCAAAACGCUUCACUGAUUUCUGCGUUGG___CAGCGGCAACUCG_CGUAAUGCAAUGGUUGCUGCUGCCACCGCAGC_____GCAUUUGAAGUUGCUUC (((((......(((((.............))))).......((((((.......................)))))).................)))))......... ( -9.96 = -9.23 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:54 2011