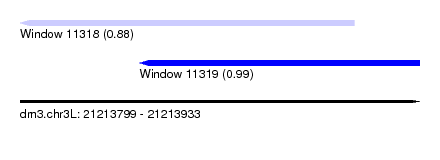
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,213,799 – 21,213,933 |
| Length | 134 |
| Max. P | 0.990625 |

| Location | 21,213,799 – 21,213,911 |
|---|---|
| Length | 112 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
| Shannon entropy | 0.33140 |
| G+C content | 0.48088 |
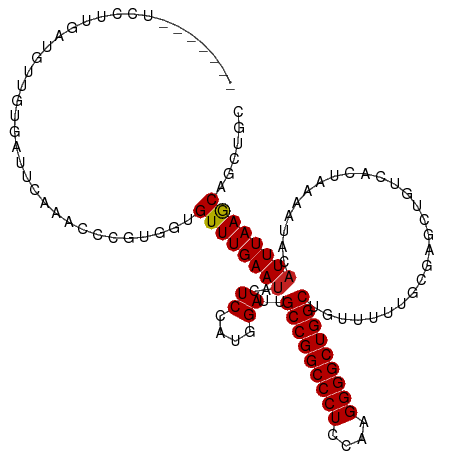
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -23.98 |
| Energy contribution | -23.87 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
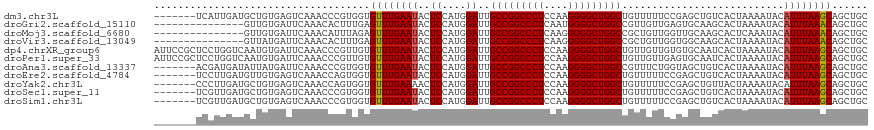
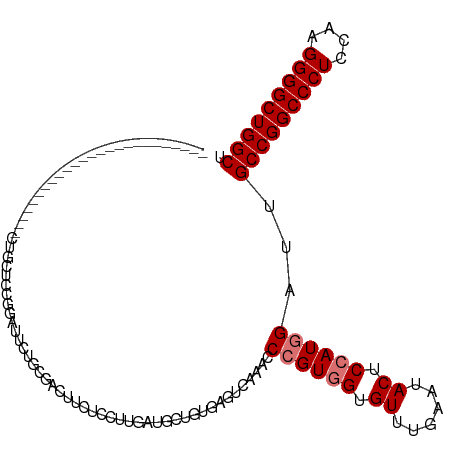
>dm3.chr3L 21213799 112 - 24543557 -------UCAUUGAUGCUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUAAGCAGCUGC -------...(((((.......)))))..((((((.((......)).))))))...(((((((((....)))))))))..........((((((...((((.....)))).)))))).. ( -37.70, z-score = -2.08, R) >droGri2.scaffold_15110 12515893 104 + 24565398 ---------------GUUGUGAUUCAAACACUUUGAGUUUGAAUACUCCAUGGAUUGCCGGCCCUCAAUGGGGCUGGCCGUUGUUGAGUGCAAGCACUAAAAUACAUUUAAACAGCUGC ---------------(((((.((((((((.......))))))))((((.(..(...(((((((((....)))))))))..)..).))))......................)))))... ( -34.50, z-score = -2.58, R) >droMoj3.scaffold_6680 8162832 104 - 24764193 ---------------GUUGUGAUUCAAACAUUUAGAGUUUGAAUACUCCAUGGAUUGCCGGCCCUCAAGGGGGCUGGCCGCUGUUGGUUGCAAGCACUCAAAUACAUUUAAACAGCUGC ---------------(((((.((((((((.......)))))))).....(((....(((((((((....))))))))).(((((.....)).))).........)))....)))))... ( -34.20, z-score = -1.87, R) >droVir3.scaffold_13049 20450490 104 - 25233164 ---------------GUUAUGAUUCAAACACUUUGAGUUUGAAUACUCCAUGGAUUGCCGGCCCUCAAGGGGGCUGGCCGCUGUUGGUGGCAAGCACUAAAAUACAUUUAAACAGCUGC ---------------.(((((((((((((.......))))))))....)))))...(((((((((....))))))))).((((((((((.....)))))............)))))... ( -34.80, z-score = -2.23, R) >dp4.chrXR_group6 12583848 119 - 13314419 AUUCCGCUCCUGGUCAAUGUGAUUCAAACCCGUUGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUGUUGUGUGCAAUCACUAAAAUACAUUUAAGCAGCUGC .....((((((((.....(((.(((((((.......)))))))))).))).)))..(((((((((....)))))))))((((..(((((............)))))....))))))... ( -35.90, z-score = -1.47, R) >droPer1.super_33 604792 119 - 967471 AUUCCGCUCCUGGUCAAUGUGAUUCAAACCCGUUGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUGUUGAGUGCAAUCACUAAAAUACAUUUAAGCAGCUGC .....(((.((....((((((((((((((.......))))))))((((.(..(...(((((((((....)))))))))..)..).)))).............))))))..)).)))... ( -38.20, z-score = -2.09, R) >droAna3.scaffold_13337 14807893 112 + 23293914 -------ACGAUGAUAUUAUGAUUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCCGUUUCUGGUAGCUGUCACUAAAAUACAUUUAAGCAGCUGC -------..............(((((((((....).))))))))...(((..(((.(((((((((....))))))))).)))..)))(((((((...((((.....)))).))))))). ( -38.90, z-score = -2.86, R) >droEre2.scaffold_4784 20898109 112 - 25762168 -------UCCUUGAUGUUGUGAGUCAAACCAGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUAAGCAGCUGC -------......(((..(((..(((((((....).)))))).)))..)))(((..(((((((((....)))))))))......))).((((((...((((.....)))).)))))).. ( -34.80, z-score = -1.30, R) >droYak2.chr3L 4885616 112 + 24197627 -------CCCUUGAUGCUGUGAGUCAAACCAGUGGUGUUUGAAAACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUUACUAAAAUACAUUUAAGCAGCUGC -------......(((..((...(((((((....).))))))..))..)))(((..(((((((((....)))))))))......))).(((((((..((((.....))))))))))).. ( -36.10, z-score = -1.63, R) >droSec1.super_11 1167145 112 - 2888827 -------UCGUUGAUGCUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUAAGCAGCUGC -------(((..((.((............((((((.((......)).))))))...(((((((((....))))))))).)).))..)))(((((...((((.....)))).)))))... ( -37.90, z-score = -2.06, R) >droSim1.chr3L 20572872 112 - 22553184 -------UCGUUGAUGCUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUCCGAGCUGUCACUAAAAUACAUUUAAGCAGCUGC -------(((..((.((............((((((.((......)).))))))...(((((((((....))))))))).)).))..)))(((((...((((.....)))).)))))... ( -37.90, z-score = -2.06, R) >consensus _______UCCUUGAUGUUGUGAUUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCUGUUUUUGCGAGCUGUCACUAAAAUACAUUUAAGCAGCUGC ....................................((((((((..((....))..(((((((((....)))))))))...........................))))))))...... (-23.98 = -23.87 + -0.11)
| Location | 21,213,839 – 21,213,933 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.46 |
| Shannon entropy | 0.43834 |
| G+C content | 0.54361 |
| Mean single sequence MFE | -36.09 |
| Consensus MFE | -25.76 |
| Energy contribution | -26.26 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21213839 94 - 24543557 -------------------------CUGCUCCGGAUUCUGUGAGUUCUCAUUGAUGCUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCU -------------------------..(((((((..((.((((....)))).))..))).)))).....((((((.((......)).))))))...(((((((((....))))))))). ( -40.00, z-score = -3.78, R) >dp4.chrXR_group6 12583888 119 - 13314419 GUUGGUGCUGUUCCCGAUCCCGAUUCCGAUCCCGAUUUCGAUUCCGCUCCUGGUCAAUGUGAUUCAAACCCGUUGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCU ...((.((.......((((........)))).((....)).....)).)).((((.(((..((((((((.......))))))))....))).))))(((((((((....))))))))). ( -35.40, z-score = -0.55, R) >droPer1.super_33 604832 119 - 967471 GUUGGUGCUGUUCCCGAUCCCGAUUCCGAUCCCGAUUUCGAUUCCGCUCCUGGUCAAUGUGAUUCAAACCCGUUGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCU ...((.((.......((((........)))).((....)).....)).)).((((.(((..((((((((.......))))))))....))).))))(((((((((....))))))))). ( -35.40, z-score = -0.55, R) >droAna3.scaffold_13337 14807933 104 + 23293914 ------------GCUGAUGAUGAUUCUGCUCCGGAUCCUCC---UUGACGAUGAUAUUAUGAUUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCC ------------(..((.((.(((((......))))).)).---))..)....................((((((.((......)).))))))...(((((((((....))))))))). ( -33.70, z-score = -1.80, R) >droEre2.scaffold_4784 20898149 94 - 25762168 -------------------------CUGCUCCGGAUUGUGCGACUUCUCCUUGAUGUUGUGAGUCAAACCAGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCU -------------------------....(((((.(((.(((((.((.....)).)))))....))).)).((((.((......)).)))))))..(((((((((....))))))))). ( -35.30, z-score = -2.31, R) >droYak2.chr3L 4885656 91 + 24197627 ----------------------------CUCCGGAUUCUGCGACUUUCCCUUGAUGCUGUGAGUCAAACCAGUGGUGUUUGAAAACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCU ----------------------------.(((.(((((.(((.(........).)))...)))))......((((.(((....))).)))))))..(((((((((....))))))))). ( -33.80, z-score = -2.09, R) >droSec1.super_11 1167185 91 - 2888827 ----------------------------CUCCGGAUUCUGUGAGUUCUCGUUGAUGCUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCU ----------------------------((((((..((..(((....)))..))..))).)))......((((((.((......)).))))))...(((((((((....))))))))). ( -36.70, z-score = -3.09, R) >droSim1.chr3L 20572912 94 - 22553184 -------------------------CUGCUCCGGAUUCUGUGAGUUCUCGUUGAUGCUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCU -------------------------..(((((((..((..(((....)))..))..))).)))).....((((((.((......)).))))))...(((((((((....))))))))). ( -38.40, z-score = -3.30, R) >consensus _________________________CUGCUCCGGAUUCUGCGACUUCUCCUUGAUGCUGUGAGUCAAACCCGUGGUGUUUGAAUACUCCAUGGAUUGCCGGCCCUCCAAGGGGCUGGCU .....................................................................((((((.((......)).))))))...(((((((((....))))))))). (-25.76 = -26.26 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:51 2011