| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,209,359 – 21,209,510 |
| Length | 151 |
| Max. P | 0.993459 |

| Location | 21,209,359 – 21,209,510 |
|---|---|
| Length | 151 |
| Sequences | 6 |
| Columns | 157 |
| Reading direction | forward |
| Mean pairwise identity | 63.85 |
| Shannon entropy | 0.63915 |
| G+C content | 0.35855 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -15.67 |
| Energy contribution | -15.20 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
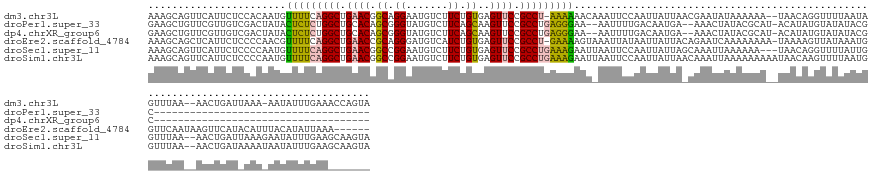
>dm3.chr3L 21209359 151 + 24543557 AAAGCAGUUCAUUCUCCACAAUGUUUUCAGGCUGAACGGCAGGAAUGUCUUCUGUGAGUUCCGCCU-AAAAAACAAAUUCCAAUUAUUAACGAAUAUAAAAAA--UAACAGGUUUUAAUAGUUUAA--AACUGAUUAAA-AAUAUUUGAAACCAGUA ...((................((((((.((((.((((.((((((.....))))))..)))).))))-..))))))...............(((((((....((--(....((((((((....))))--)))).)))...-.)))))))......)). ( -30.00, z-score = -1.99, R) >droPer1.super_33 600446 116 + 967471 GAAGCUGUUCGUUGUCGACUAUACUCUCUGGCUGCACAGCGGGUAUGUCUUCAGCAAGUUCCGCCUGAGGGAA--AAUUUUGACAAUGA--AAACUAUACGCAU-ACAUAUGUAUAUACGC------------------------------------ ...((..(((((((((((.(((((((.(((......))).)))))))((((((((.......).)))))))..--....))))))))))--)...((((..(((-....)))..)))).))------------------------------------ ( -33.30, z-score = -2.03, R) >dp4.chrXR_group6 12579512 116 + 13314419 GAAGCUGUUCGUUGUCGACUAUACUCUCUGGCUGCACAGCGGGUAUGUCUUCAGCAAGUUCCGCCUGAGGGAA--AAUUUUGACAAUGA--AAACUAUACGCAU-ACAUAUGUAUAUACGC------------------------------------ ...((..(((((((((((.(((((((.(((......))).)))))))((((((((.......).)))))))..--....))))))))))--)...((((..(((-....)))..)))).))------------------------------------ ( -33.30, z-score = -2.03, R) >droEre2.scaffold_4784 20893793 149 + 25762168 AAAGCAGCUCAUUCUCCCCAACGUUUUCAGGCUGAACCGCAGGGAUGUCAUCUGUGAGUUCCGCCU-GAAAAGUAAAUUAUAAUUAUUACAGAAUCAAAAAAAA-UAAAAGUUAUAAAUGGUUCAAUAAGUUCAUACAUUUACAUAUUAAA------ .......................(((((((((.((((((((((.......)))))).)))).))))-)))))((((((........(((..((((((.....((-(....))).....))))))..)))........))))))........------ ( -32.69, z-score = -3.23, R) >droSec1.super_11 1162887 152 + 2888827 AAAGCAGUUCAUUCUCCCCAAUGUUUUCAGGCUGAACGGCCGGAAUGUCUUCUGUGAGUUCCGCCUGAAAGAAUUAAUUCCAAUUAUUAGCAAAUUAAAAAA---UAACAGGUUUUAUUGGUUUAA--AACUGAUUAAAGAAUAUUUGAAGCAAGUA ...((..............(((.(((((((((.((((..((((((....))))).).)))).))))))))).)))...............(((((.......---(((..(((((((......)))--))))..)))......)))))..))..... ( -31.82, z-score = -0.91, R) >droSim1.chr3L 20568631 155 + 22553184 AAAGCAGUUCAUUCUCCCCAAUGUUUUCAGGCUGAACGGCCGGAAUGUCUUCUGUGAGUUCCGCCUGAAAGAAUUAAUUCCAAUUAUUAACAAAUUAAAAAAAAAUAACAAGUUUUAAUGGUUUAA--AACUGAUAAAAUAAUAUUUGAAGCAAGUA ...((..............(((.(((((((((.((((..((((((....))))).).)))).))))))))).))).....(((.(((((.....(((........)))..((((((((....))))--)))).......))))).)))..))..... ( -31.00, z-score = -1.76, R) >consensus AAAGCAGUUCAUUCUCCACAAUGUUUUCAGGCUGAACGGCAGGAAUGUCUUCUGUGAGUUCCGCCUGAAAGAAU_AAUUCCAAUUAUUA_CAAAUUAAAAAAA__UAACAGGUAUAAAUGGUUUAA__AACUGAU_AAA_AAUAUUUGAA_______ .......................(((((((((.((((.((.((.......)).))..)))).)))))))))...................................................................................... (-15.67 = -15.20 + -0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:49 2011