
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,169,448 – 21,169,538 |
| Length | 90 |
| Max. P | 0.696636 |

| Location | 21,169,448 – 21,169,538 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 69.69 |
| Shannon entropy | 0.63238 |
| G+C content | 0.40537 |
| Mean single sequence MFE | -20.61 |
| Consensus MFE | -8.42 |
| Energy contribution | -8.58 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.696636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
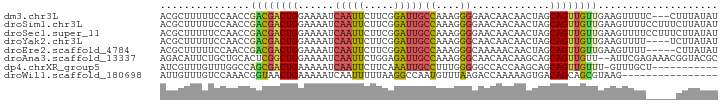
>dm3.chr3L 21169448 90 + 24543557 ACGCUUUUUCCAACCGACGACUGGAAAAUCAAUUCUUCGGAUUGCCAAAGGGGAACAACAACUAGCAGUUGUUGAAGUUUUC---CUUUAUAU ..((.(((((((.........))))))).....((....))..)).(((((..(((..((((........))))..)))..)---)))).... ( -23.30, z-score = -2.26, R) >droSim1.chr3L 20521746 93 + 22553184 ACGCUUUUUCCAACCGACGACUGGAAAAUCAAUUCUUCGGAUUGCCAAAGGGGAACAACAACUAGCAGUUGUUGAAGUUUUCCUUUCUUAUAU ..((.(((((((.........))))))).....((....))..)).(((((..(((..((((........))))..)))..)))))....... ( -22.70, z-score = -1.88, R) >droSec1.super_11 1122762 93 + 2888827 ACGCUUUUUCCAACCGACGACUGGAAAAUCAAUUCUUCGGAUUGCCAAAGGGGAACAACAACUAGCAGUUGUUGAAGUUUUCCUUUCUUAUAU ..((.(((((((.........))))))).....((....))..)).(((((..(((..((((........))))..)))..)))))....... ( -22.70, z-score = -1.88, R) >droYak2.chr3L 4839569 89 - 24197627 ACGCUUUUUCCAACCGACGACUGGAAAAUCAAUUCUUCGGAUUGCCAAAGGGCAACAACAACUAGCAGUUGUUGAAGUUUU----UCUUAUAU ..(((((((.(((((((.((...(.....)...)).)))).)))..)))))))..(((((((.....))))))).......----........ ( -20.90, z-score = -2.00, R) >droEre2.scaffold_4784 20854045 88 + 25762168 ACGCUUUUUCCAACCGACGACUGGAAAAUCAAUUCUUCGGAUUGCCAAAGGGCAAAAACAACUAGCAGUUGUUGAAGUUUU-----CUUAUAU ..............(((((((((..........((....))(((((....)))))..........))))))))).......-----....... ( -20.30, z-score = -1.70, R) >droAna3.scaffold_13337 14763055 91 - 23293914 AGACAUUCUGCUGCACUCGGCUGGAAAAUCAAUUCUGGAGAUUGCCAAAGGGCAACAACAAGCAGCAGUUGUU--AUUCGAGAAACGGUACGC .((((..(((((((..((..(..(((......)))..).))(((((....)))))......))))))).))))--.................. ( -27.00, z-score = -1.93, R) >dp4.chrXR_group5 641610 81 + 740492 AUCGUUUGUUUGGCCAGCGACUGAAAAAUCAAUUCUUCAAAUUGCCUUUGGGGGCCACCAAGCAGCAGUUGUUU-GUUUGCU----------- ...(((((..(((((.....(..((....(((((.....)))))..))..).))))).)))))(((((......-..)))))----------- ( -19.20, z-score = 0.39, R) >droWil1.scaffold_180698 5567796 77 - 11422946 AUUGUUUGUCCAAACGGUAACUGAAAAAUCAAUUUUUAAGGCCAAUGUUUAAGACCAAAAAGUGACAGCAGCGUAAG---------------- .(((((.((((....(((...((((((.....))))))..)))...(((...)))......).))))))))......---------------- ( -8.80, z-score = 0.96, R) >consensus ACGCUUUUUCCAACCGACGACUGGAAAAUCAAUUCUUCGGAUUGCCAAAGGGGAACAACAACUAGCAGUUGUUGAAGUUUU_____CUUAUAU ...............((((((((......(((((.....)))))((....)).............)))))))).................... ( -8.42 = -8.58 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:41 2011