| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,116,269 – 21,116,364 |
| Length | 95 |
| Max. P | 0.745547 |

| Location | 21,116,269 – 21,116,364 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.56549 |
| G+C content | 0.51091 |
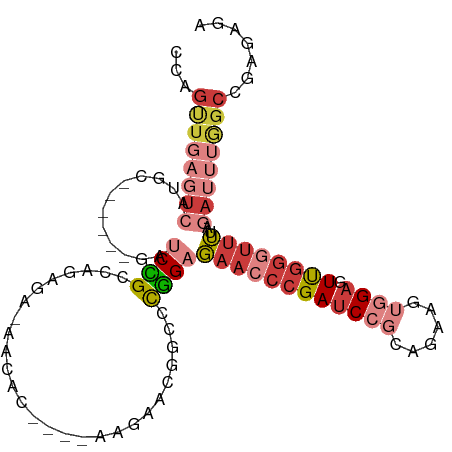
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -11.09 |
| Energy contribution | -13.22 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
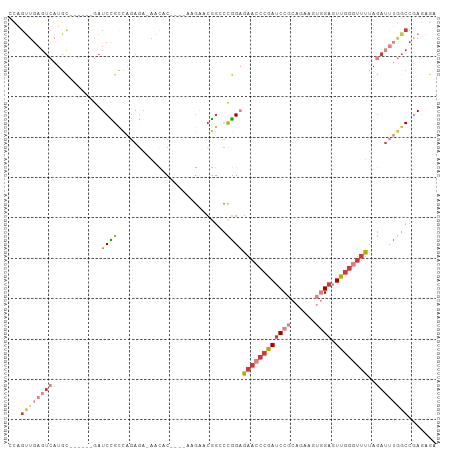
>dm3.chr3L 21116269 95 - 24543557 CCAGUUGAGUCAUGC------GAUCCGCCAGAGA-AACAC----AAGAACGGCCCGGAGAACCCGAUCCGCAGAAGUGGAGUUGGGUUUUAGAUUUGGCCGAGAGA ...(((...((..((------(...)))..))..-)))..----.....(((((...((((((((((((((....))))).)))))))))......)))))..... ( -30.60, z-score = -2.49, R) >droSim1.chr3L 20471627 95 - 22553184 CCAGUUGAGUCAUGC------GAUCCGUCAGAGA-AACAC----AAGAACGGCCCGGAGAACCCGAUCCGCAGAAAUGGAGUUGGGUUUUAGAUUUGGCCGAGAGA ...((..((((...(------(..(((((.....-.....----..).))))..)).(((((((((..(.((....)))..))))))))).))))..))....... ( -26.90, z-score = -1.24, R) >droSec1.super_11 1074031 95 - 2888827 CCAGUUGAGUCAUGC------GAUCCGCCAGAGA-AACAC----AAGAACGGCCCGGUGAACCCGAUCCGCAGAAAUGGAGUUGGGUUUUAGAUUUGGCCGAGAGA ...(((...((..((------(...)))..))..-)))..----.....(((((..((((((((((..(.((....)))..))))))))...))..)))))..... ( -27.30, z-score = -1.22, R) >droYak2.chr3L 4790666 95 + 24197627 CCAGUUGAGUCAUGC------GAUCCGCCAGAGA-AACAC----AAGAACGGGCCGGAGAACCCGAUCCGCAGAAGUGGAGUUGGGUUUUAGAUUUGGCCAAGAGA ...((..((((...(------(..(((.......-.....----.....)))..)).((((((((((((((....))))).))))))))).))))..))....... ( -31.03, z-score = -2.52, R) >droEre2.scaffold_4784 20805361 95 - 25762168 CCAGUUGAGUCAUGC------GAUCCGCCAGAGA-AACAC----AAGAACGGGCCGGAGAACCCGAUCCGCAGAAGUGGAGUUGGGUUUGAGAUUUGGCCAAGAGA ...((..((((....------..((((((.....-.....----.......)).))))(((((((((((((....))))).))))))))..))))..))....... ( -31.03, z-score = -2.33, R) >droAna3.scaffold_13337 14716557 97 + 23293914 AUGGCUGAGUCAUGUUGCAUGUAUCUGCAGAAAACAAAACGGCGAAGAACAGGUCAGAGAACCCGAUCC---GAAGUCGACUUGG--CUUAAA---AGCCGAAGA- .(((((((((((((((...(((....)))...))))...((((...((.(.(((......))).).)).---...))))...)))--)))...---)))))....- ( -23.50, z-score = -0.40, R) >droWil1.scaffold_181017 99633 81 - 939937 ---------------------GAUCUGGUUGAGU-CAUAUAAAAAAAGAAGACUUGGCCAA-CAGAGCCAGCUAAGAAGAGUCA--UUGUUGUUGUGGCAGAAAGA ---------------------..((((((..(((-(..............))))..)))..-....((((..(((.((......--)).)))...))))))).... ( -18.64, z-score = -1.32, R) >consensus CCAGUUGAGUCAUGC______GAUCCGCCAGAGA_AACAC____AAGAACGGCCCGGAGAACCCGAUCCGCAGAAGUGGAGUUGGGUUUUAGAUUUGGCCGAGAGA ...((..((((............((((...........................))))((((((((((((......)))).))))))))..))))..))....... (-11.09 = -13.22 + 2.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:38 2011