| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,079,445 – 21,079,545 |
| Length | 100 |
| Max. P | 0.710469 |

| Location | 21,079,445 – 21,079,545 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.41085 |
| G+C content | 0.43732 |
| Mean single sequence MFE | -17.04 |
| Consensus MFE | -7.43 |
| Energy contribution | -8.32 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
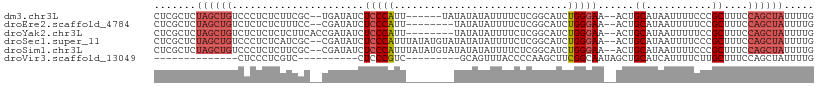
>dm3.chr3L 21079445 100 + 24543557 CUCGCUCUAGCUGUCCCUCUCUUCGC--UGAUAUCUCCCAUU------UAUAUAUAUUUUCUCGGCAUCUGGGAA--ACUGCAUAAUUUUCCCGCUUUCCAGCUAUUUUG .......((((((...........((--(((...........------.............)))))....(((((--(.(.....).))))))......))))))..... ( -18.26, z-score = -2.27, R) >droEre2.scaffold_4784 20767459 98 + 25762168 CUCGCUCUAGCUGUCUCUCUCUUUCC--CGAUAUCUCCCAUU--------UAUAUAUUUUCUCGGCAUCUGGGAA--ACUGCAUAAUUUUUCCGCUUUCCAGCUAUUUUG .......((((((........(((((--((((..(.......--------.............)..))).)))))--)..((...........))....))))))..... ( -14.75, z-score = -1.44, R) >droYak2.chr3L 4752175 100 - 24197627 CUCGCUCUAGCUGUCUCUCUCUCUUCACCGAUAUCUCCCAUU--------UAUAUAUUUUCUCGGCAUCUGGGAA--ACUGCAUAAUUUUUCCGCUUUCCAGCUAUUUUG .......((((((..............((((...........--------...........)))).....(((((--(.(.....).))))))......))))))..... ( -13.15, z-score = -0.99, R) >droSec1.super_11 1037151 106 + 2888827 CUCGCUCUAGCUGUCCCUCUCAUCGC--CGAUAUCUCCCAUUUAUAUGUAUAUAUAUUUUCUCGGCAUCUGGGAA--ACUGCAUAAUUUUCCCGCUUUCCAGCUAUUUUG .......((((((...........((--(((...........(((((....))))).....)))))....(((((--(.(.....).))))))......))))))..... ( -20.29, z-score = -2.42, R) >droSim1.chr3L 20434526 106 + 22553184 CUCGCUCUAGCUGUCCCUCUCUUCGC--CGAUAUCUCCCAUUUAUAUGUAUAUAUAUUUUCUCGGCAUCUGGGAA--ACUGCAUAAUUUUCCCGCUUUCCAGCUAUUUUG .......((((((...........((--(((...........(((((....))))).....)))))....(((((--(.(.....).))))))......))))))..... ( -20.29, z-score = -2.58, R) >droVir3.scaffold_13049 9970655 77 - 25233164 --------------CUCCCUCGUC----------CUCCCGUC---------GCAGUUUACCCCAAGCUUCGGCAAUAGCUGCAUCAUUUUCUUGCUUUCCAGCUAUUUUG --------------..........----------.....(((---------(.(((((.....))))).))))((((((((...((......)).....))))))))... ( -15.50, z-score = -3.55, R) >consensus CUCGCUCUAGCUGUCCCUCUCUUCGC__CGAUAUCUCCCAUU________UAUAUAUUUUCUCGGCAUCUGGGAA__ACUGCAUAAUUUUCCCGCUUUCCAGCUAUUUUG .......((((((......................(((((.............................)))))......((...........))....))))))..... ( -7.43 = -8.32 + 0.89)
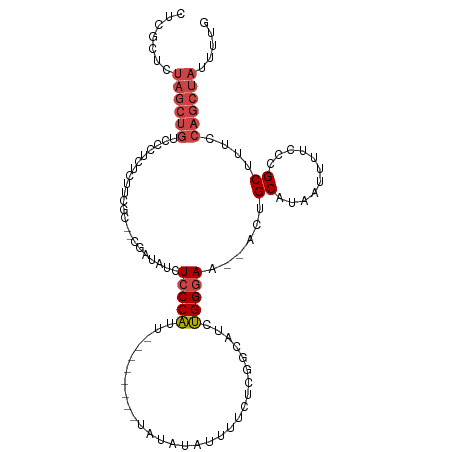

| Location | 21,079,445 – 21,079,545 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.41085 |
| G+C content | 0.43732 |
| Mean single sequence MFE | -20.00 |
| Consensus MFE | -11.33 |
| Energy contribution | -12.08 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21079445 100 - 24543557 CAAAAUAGCUGGAAAGCGGGAAAAUUAUGCAGU--UUCCCAGAUGCCGAGAAAAUAUAUAUA------AAUGGGAGAUAUCA--GCGAAGAGAGGGACAGCUAGAGCGAG .....((((((....(((.........))).((--((((((.....................------..))))))))....--.............))))))....... ( -18.60, z-score = -0.86, R) >droEre2.scaffold_4784 20767459 98 - 25762168 CAAAAUAGCUGGAAAGCGGAAAAAUUAUGCAGU--UUCCCAGAUGCCGAGAAAAUAUAUA--------AAUGGGAGAUAUCG--GGAAAGAGAGAGACAGCUAGAGCGAG .....((((((....(((.........)))..(--(((((.((((((.(...........--------..).))...)))))--)))))........))))))....... ( -19.32, z-score = -1.82, R) >droYak2.chr3L 4752175 100 + 24197627 CAAAAUAGCUGGAAAGCGGAAAAAUUAUGCAGU--UUCCCAGAUGCCGAGAAAAUAUAUA--------AAUGGGAGAUAUCGGUGAAGAGAGAGAGACAGCUAGAGCGAG .....((((((....(((.........))).((--((((((...................--------..))))))))...................))))))....... ( -18.70, z-score = -1.65, R) >droSec1.super_11 1037151 106 - 2888827 CAAAAUAGCUGGAAAGCGGGAAAAUUAUGCAGU--UUCCCAGAUGCCGAGAAAAUAUAUAUACAUAUAAAUGGGAGAUAUCG--GCGAUGAGAGGGACAGCUAGAGCGAG .....((((((......((((((.........)--)))))...((((((.....(((.(((....))).))).......)))--)))..........))))))....... ( -23.80, z-score = -2.07, R) >droSim1.chr3L 20434526 106 - 22553184 CAAAAUAGCUGGAAAGCGGGAAAAUUAUGCAGU--UUCCCAGAUGCCGAGAAAAUAUAUAUACAUAUAAAUGGGAGAUAUCG--GCGAAGAGAGGGACAGCUAGAGCGAG .....((((((......((((((.........)--)))))...((((((.....(((.(((....))).))).......)))--)))..........))))))....... ( -23.80, z-score = -2.30, R) >droVir3.scaffold_13049 9970655 77 + 25233164 CAAAAUAGCUGGAAAGCAAGAAAAUGAUGCAGCUAUUGCCGAAGCUUGGGGUAAACUGC---------GACGGGAG----------GACGAGGGAG-------------- ...((((((((.(...((......)).).)))))))).(((..((..((......))))---------..)))...----------..........-------------- ( -15.80, z-score = -0.69, R) >consensus CAAAAUAGCUGGAAAGCGGGAAAAUUAUGCAGU__UUCCCAGAUGCCGAGAAAAUAUAUA________AAUGGGAGAUAUCG__GCGAAGAGAGAGACAGCUAGAGCGAG .....((((((....(((.........))).....((((((.............................)))))).....................))))))....... (-11.33 = -12.08 + 0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:35 2011