
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,967,062 – 5,967,162 |
| Length | 100 |
| Max. P | 0.865425 |

| Location | 5,967,062 – 5,967,162 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 58.96 |
| Shannon entropy | 0.79653 |
| G+C content | 0.55170 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -13.62 |
| Energy contribution | -12.78 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.865425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
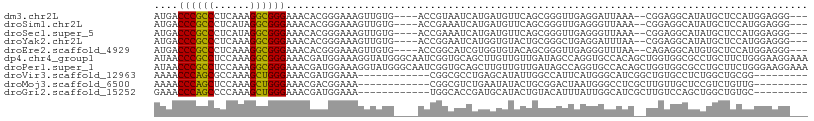
>dm3.chr2L 5967062 100 + 23011544 AUGACCCGCCCUCAAAGGCGGGAAACACGGGAAAGUUGUG----ACCGUAAUCAUGAUGUUCAGCGGGUUGAGGAUUAAA--CGGAGGCAUAUGCUCCAUGGAGGG--- ....((((((......))))))...(((((.....)))))----.((((.....((((.(((((....))))).))))..--.((((.(....)))))))))....--- ( -30.60, z-score = -0.59, R) >droSim1.chr2L 5755037 100 + 22036055 AUGACCCGCCCUCAUAGGCGGGAAACACGGGAAAGUUGUG----ACCGAAAUCAUGAUGUUCAGCGGGUUGAGGGUUAAA--CGGAGGCAUAUGCUCCAUGGAGGG--- ....((((((((((...((.(....)....(((.((..((----(......)))..)).))).))....)))))))....--.((((.(....))))).....)))--- ( -31.50, z-score = -0.63, R) >droSec1.super_5 4038020 100 + 5866729 AUGACCCGCCCUCAUAGGCGGGAAACACGGGAAAGUUGUG----ACCGAAAUCAUGAUGUUCAGCGGGUUGAGGGUUAAA--CGGAGGCAUAUGCUCCAUGGAGGG--- ....((((((((((...((.(....)....(((.((..((----(......)))..)).))).))....)))))))....--.((((.(....))))).....)))--- ( -31.50, z-score = -0.63, R) >droYak2.chr2L 9096599 100 - 22324452 AUGACCCGCCCUCAAAGGCGGGAAACACGGGAAAGUUGUG----ACCGGAAUCAUGGUGUACUGCGGGCUGAGGAUUUAA--CGGAGGCAUAUGCUCCAUGGAGGG--- ....((((((......))))))...(((((.....)))))----.((....((((((.(((.(((...(((.........--)))..)))..))).)))))).)).--- ( -30.00, z-score = 0.12, R) >droEre2.scaffold_4929 6057372 100 + 26641161 AUGACCCGCCCUCAAAGGCGGGAAACACGGGAAAGUUGUG----ACCGGCAUCGUGGUGUACAGCGGGUUGAGGGUUUAA--CAGAGGCAUGUGCUCCAUGGAGGG--- ....((((((......))))))...(((((.....)))))----.((....((((((.(((((((..(((((....))))--)....)).))))).)))))).)).--- ( -34.80, z-score = -0.65, R) >dp4.chr4_group1 4960429 109 - 5278887 AUAACCCGCCUCCAAAGGCGGGAAACGAUGGAAAGGUAUGGGCAAUCGGUGCAGCUUGUUGUUGAUAGCCAGGUGCCACAGCUGGUGGCGCCUGCUUCUGGGAAGGAAA ....(((((((....))))))).......((((.(.(((.((((((.((.....)).)))))).))).)((((((((((.....)))))))))).)))).......... ( -43.70, z-score = -1.60, R) >droPer1.super_1 9968913 109 - 10282868 AUAACCCGCCUCCAAAGGCGGGAAACGAUGGAAAGGUAUGGGCAAUCGGUGCAGCUUGUUGUUGAUAGCCAGGUGCCACAGCUGGUGGCGCCUGCUUCUGGGAAGGAAA ....(((((((....))))))).......((((.(.(((.((((((.((.....)).)))))).))).)((((((((((.....)))))))))).)))).......... ( -43.70, z-score = -1.60, R) >droVir3.scaffold_12963 3251851 88 + 20206255 AAAACCCAGCGCCAAAGCUGGGAAACGAUGGAAA------------CGGCGCCUGAGCAUAUUGGCCAUUCAUGGGCAUCGGCUGUGCCUCUGGCUGCGG--------- ....((((((......))))))......((....------------))(((((.(((((((.((.(((....))).)).....)))).))).))).))..--------- ( -32.60, z-score = -0.62, R) >droMoj3.scaffold_6500 13615475 88 + 32352404 AAAACCCAGCUCCAAAGCUGGGAAACGACGGAAA------------CGGCGUCUGAAUAUACUGCGGACUAAUGGGCCUCGCUUGUUGCUCCGUCUGUUG--------- ....(((((((....)))))))...(((((((..------------.((((((((.........))))).((..(((...)))..)))))...)))))))--------- ( -32.20, z-score = -3.33, R) >droGri2.scaffold_15252 2836769 88 + 17193109 GAAACCCAGCCCCAAAGCUGGGAAACGAUGGAAA------------UGGCACCGAUGCAUACUGUACAUUUAUUGGCAUCGCUUGUCCAGCUGGCUGUGC--------- ......(((((....(((((((...(((((....------------..(((....)))........((.....)).)))))....))))))))))))...--------- ( -25.90, z-score = -0.65, R) >consensus AUAACCCGCCCCCAAAGGCGGGAAACAAGGGAAAGUUGUG____ACCGGAAUCAUGAUGUACUGCGGGUUGAGGGUCAAA__CGGAGGCACAUGCUCCAUGGAGGG___ ....((((((......))))))....................................................................................... (-13.62 = -12.78 + -0.84)


| Location | 5,967,062 – 5,967,162 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 58.96 |
| Shannon entropy | 0.79653 |
| G+C content | 0.55170 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -13.65 |
| Energy contribution | -12.73 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.41 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5967062 100 - 23011544 ---CCCUCCAUGGAGCAUAUGCCUCCG--UUUAAUCCUCAACCCGCUGAACAUCAUGAUUACGGU----CACAACUUUCCCGUGUUUCCCGCCUUUGAGGGCGGGUCAU ---........(((((....).))))(--((((.............)))))...((((.(((((.----..........)))))...(((((((....))))))))))) ( -24.62, z-score = -0.41, R) >droSim1.chr2L 5755037 100 - 22036055 ---CCCUCCAUGGAGCAUAUGCCUCCG--UUUAACCCUCAACCCGCUGAACAUCAUGAUUUCGGU----CACAACUUUCCCGUGUUUCCCGCCUAUGAGGGCGGGUCAU ---(((...(((((((....).)))))--)....((((((...((..((((((...((....(((----....))).))..))))))..))....)))))).))).... ( -24.10, z-score = -0.17, R) >droSec1.super_5 4038020 100 - 5866729 ---CCCUCCAUGGAGCAUAUGCCUCCG--UUUAACCCUCAACCCGCUGAACAUCAUGAUUUCGGU----CACAACUUUCCCGUGUUUCCCGCCUAUGAGGGCGGGUCAU ---(((...(((((((....).)))))--)....((((((...((..((((((...((....(((----....))).))..))))))..))....)))))).))).... ( -24.10, z-score = -0.17, R) >droYak2.chr2L 9096599 100 + 22324452 ---CCCUCCAUGGAGCAUAUGCCUCCG--UUAAAUCCUCAGCCCGCAGUACACCAUGAUUCCGGU----CACAACUUUCCCGUGUUUCCCGCCUUUGAGGGCGGGUCAU ---......(((((((....).)))))--)..........(.(((.(((........))).))).----)...........(((...(((((((....))))))).))) ( -24.20, z-score = -0.00, R) >droEre2.scaffold_4929 6057372 100 - 26641161 ---CCCUCCAUGGAGCACAUGCCUCUG--UUAAACCCUCAACCCGCUGUACACCACGAUGCCGGU----CACAACUUUCCCGUGUUUCCCGCCUUUGAGGGCGGGUCAU ---.....(((((.((....)).....--...........(((.((.............)).)))----..........)))))...(((((((....))))))).... ( -23.62, z-score = 0.56, R) >dp4.chr4_group1 4960429 109 + 5278887 UUUCCUUCCCAGAAGCAGGCGCCACCAGCUGUGGCACCUGGCUAUCAACAACAAGCUGCACCGAUUGCCCAUACCUUUCCAUCGUUUCCCGCCUUUGGAGGCGGGUUAU ...........((((((((.(((((.....))))).)))(((.(((................))).)))..............)))))((((((....))))))..... ( -32.69, z-score = -0.84, R) >droPer1.super_1 9968913 109 + 10282868 UUUCCUUCCCAGAAGCAGGCGCCACCAGCUGUGGCACCUGGCUAUCAACAACAAGCUGCACCGAUUGCCCAUACCUUUCCAUCGUUUCCCGCCUUUGGAGGCGGGUUAU ...........((((((((.(((((.....))))).)))(((.(((................))).)))..............)))))((((((....))))))..... ( -32.69, z-score = -0.84, R) >droVir3.scaffold_12963 3251851 88 - 20206255 ---------CCGCAGCCAGAGGCACAGCCGAUGCCCAUGAAUGGCCAAUAUGCUCAGGCGCCG------------UUUCCAUCGUUUCCCAGCUUUGGCGCUGGGUUUU ---------.....(((...))).....(((((.....(((((((......((....))))))------------))).)))))...((((((......)))))).... ( -24.90, z-score = 1.32, R) >droMoj3.scaffold_6500 13615475 88 - 32352404 ---------CAACAGACGGAGCAACAAGCGAGGCCCAUUAGUCCGCAGUAUAUUCAGACGCCG------------UUUCCGUCGUUUCCCAGCUUUGGAGCUGGGUUUU ---------.....((((((((.....(((.(((......)))))).((........))...)------------.)))))))....(((((((....))))))).... ( -29.10, z-score = -2.63, R) >droGri2.scaffold_15252 2836769 88 - 17193109 ---------GCACAGCCAGCUGGACAAGCGAUGCCAAUAAAUGUACAGUAUGCAUCGGUGCCA------------UUUCCAUCGUUUCCCAGCUUUGGGGCUGGGUUUC ---------...(((((((((((..((((((((.....((((((((.(.......).))).))------------))).)))))))).))))))....)))))...... ( -28.00, z-score = -0.90, R) >consensus ___CCCUCCAUGGAGCAUAUGCCACCG__UUUAACCCUCAACCCGCAGUACACCACGAUGCCGGU____CACAACUUUCCCGCGUUUCCCGCCUUUGAGGGCGGGUCAU .......................................................................................(((((((....))))))).... (-13.65 = -12.73 + -0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:11 2011