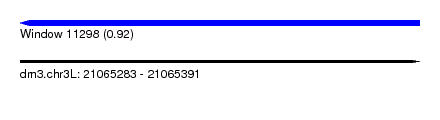
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,065,283 – 21,065,391 |
| Length | 108 |
| Max. P | 0.923875 |

| Location | 21,065,283 – 21,065,391 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 60.31 |
| Shannon entropy | 0.76327 |
| G+C content | 0.61785 |
| Mean single sequence MFE | -40.26 |
| Consensus MFE | -18.01 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923875 |
| Prediction | RNA |
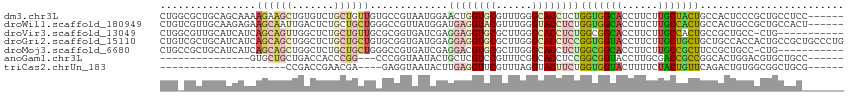
Download alignment: ClustalW | MAF
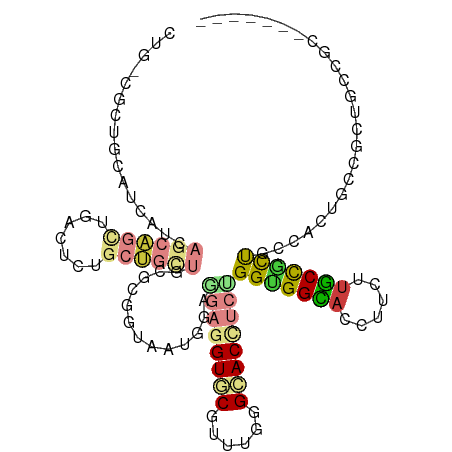
>dm3.chr3L 21065283 108 - 24543557 CUGGCGCUGCAGCAAAAGAAGCUGUGUCUGCUGUUGUGCCGUAAUGGAACUGGUGCGUUUGGGCACCUCUGGUGGCACCUUCUUGCUACUGCCACUCCCGCUGCCUCC------ ..(((((.((((((..............)))))).)))))(((.(((....(((((......)))))...(((((((............))))))).))).)))....------ ( -38.34, z-score = -0.71, R) >droWil1.scaffold_180949 1045679 108 + 6375548 CUGUCGUUGCAAGAGAAGCAAUUGACUCUGCUGCUGGGCCGUUAUGGAUGAGGUACGUUUGGGUACCUCUGGUGGCACCUUCUUGCCACUGCCACUGCCGCUGCCACU------ ..((((((((.......)))).))))...((.((..(((..........(((((((......))))))).(((((((......))))))))))...)).)).......------ ( -38.10, z-score = -0.77, R) >droVir3.scaffold_13049 9954954 102 + 25233164 CUGGCGUUGCAUCAUCAGCAGUUGGCUCUGCUGUUGCGCGGUGAUCGAGGAGGUGCGCUUGGGCACCUCUGGCGGCACCUUCUUGCCACUGCCGCUGCC-CUG----------- ..((((..(((....((((((......)))))).)))((((((..((((((((((((((.(((...))).))).))))))))))).))))))))))...-...----------- ( -48.80, z-score = -1.91, R) >droGri2.scaffold_15110 4883056 114 + 24565398 CUGUCGCUGCAUCAUCAGCAGCUGGCUCUGCUGCUGUGCGGUGAUGGAGGAGGUGCGCUUGGGCACCUCCGGUGGUACCUUCUUGCUGCUGCUGCCACCACUGCCGCUGCCCUG (((((((((((....(((((((.......)))))))))))))))))).((((((((......))))))))(((((((.(..(.....)..).)))))))............... ( -55.10, z-score = -2.24, R) >droMoj3.scaffold_6680 21584740 102 - 24764193 CUGCCGCUGCAUCAUCAGCAGCUGGCUCUGCUGCUGGGCCGUGAUCGAGGACGUGCGCUUGGGCACCUCUGGCGGCACCUUCUUGCCGCUUCCGCUGCC-CUG----------- ..((((((((.......))))).))).........((((.(((.....(((.((((......)))).)))(((((((......)))))))..))).)))-)..----------- ( -43.70, z-score = -0.68, R) >anoGam1.chr3L 23534213 90 - 41284009 ---------------GUGCUGCUGACCACCCGG---CCCGGUAAUACUGCUCGUCCGUUUCGGCACCUCCGGCGGUACCUUGCGACCGCCGGCACUGGACGUGCUGCC------ ---------------(((.(((((.((....))---..))))).))).((.(((((((....))....((((((((........))))))))....))))).))....------ ( -34.80, z-score = -0.59, R) >triCas2.chrUn_183 56953 83 + 157303 ---------------------CCGACCGAACGA----GAGGUAAUACUUGAGGUUCGUUUAGGUACUUCUGGUGGUACUUUUCUACUGUUCAGACUGUGGCGGCUGCG------ ---------------------(((.(((...((----((((((.((((.(((((((.....)).))))).)))).))))))))..((....))....)))))).....------ ( -23.00, z-score = -0.18, R) >consensus CUG_CGCUGCAUCAUCAGCAGCUGACUCUGCUGCUGCGCGGUAAUGGAGGAGGUGCGUUUGGGCACCUCUGGUGGCACCUUCUUGCCGCUGCCACUGCCGCUGCCGC_______ ................((((((.......)))))).............((((((((......))))))))(((((((......)))))))........................ (-18.01 = -18.93 + 0.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:33 2011