| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,060,933 – 21,061,027 |
| Length | 94 |
| Max. P | 0.855738 |

| Location | 21,060,933 – 21,061,027 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 63.65 |
| Shannon entropy | 0.76785 |
| G+C content | 0.57528 |
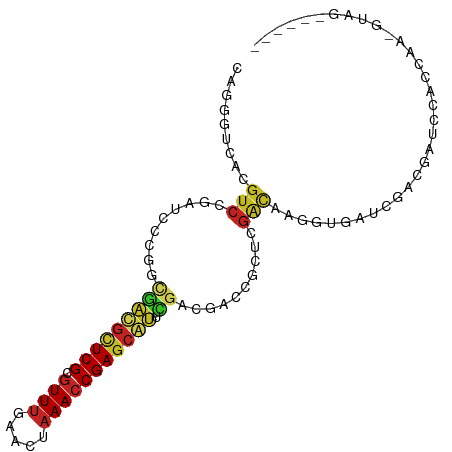
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -11.32 |
| Energy contribution | -11.05 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
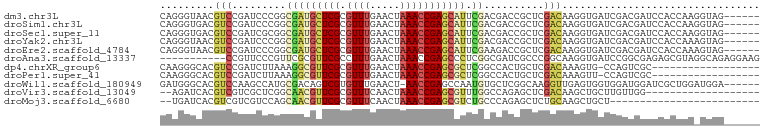
>dm3.chr3L 21060933 94 - 24543557 CAGGGUAACGUCCGAUCCCGGCGAUGCUCGCGUUUGAACUAAACCGAGCAUUCGACGACCGCUCGACAAGGUGAUCGACGAUCCACCAAGGUAG------ ..((((..((((.((((((..(((((((((.(((((...))))))))))).))).(((....)))....)).))))))))))))..........------ ( -31.50, z-score = -1.41, R) >droSim1.chr3L 20415121 94 - 22553184 CAGGGUGACGUCCGAUCCCGGCGAUGCUCGCGUUUGAACUAAACCGAGCAUUCGACGACCGCUCGACAAGGUGAUCGACGAUCCACCAAGGUAG------ ...((((.((((.((((((..(((((((((.(((((...))))))))))).))).(((....)))....)).))))))))...)))).......------ ( -33.30, z-score = -1.58, R) >droSec1.super_11 949510 94 - 2888827 CAGGGUGACGUCCGAUCGCGGCGAUGCUCGCGUUUGAACUAAACCGAGCAUUCGACGACCGCUCGACAAGGUGAUCGACGAUCCACCAAGGUAG------ ...((((.((((.((((((..(((((((((.(((((...))))))))))).))).(((....))).....))))))))))...)))).......------ ( -34.20, z-score = -1.51, R) >droYak2.chr3L 4730378 94 + 24197627 CAGGGUAACGUCCGAUCCCGGCGAUGCUCGCGUUUGAACUAAACCGAGCAUUCGACGACCGCUCGACAAGGUGAUCGACGAUCCACCAAAGUAG------ ..((((..((((.((((((..(((((((((.(((((...))))))))))).))).(((....)))....)).))))))))))))..........------ ( -31.50, z-score = -1.87, R) >droEre2.scaffold_4784 20748387 94 - 25762168 CAGGGUAACGUCCGAUCCCGGCGAUGCUCGCGUUUGAACUAAACCGAGCAUUCGAAGACCGCUCGACAAGGUGAUCGACGAUCCACCAAAGUAG------ ..((((..((((.((((((....(((((((.(((((...))))))))))))((((.......))))...)).))))))))))))..........------ ( -30.20, z-score = -1.75, R) >droAna3.scaffold_13337 14658988 89 + 23293914 -----------CCGUUCCCGUUCGCGUUCGCCUUUGAACUAAACCGAGCCCUCGGCGAUCGCCCGGCAAGGUGAUCCGGCGAGAGCGUAGGCAGAGGAAG -----------((....((...(((.((((((...........((((....)))).(((((((......))))))).)))))).)))..))....))... ( -34.10, z-score = -0.93, R) >dp4.chrXR_group6 7365436 81 - 13314419 CAAGGGCACGUCCGAUCUUAAAGGCGUUCGCGUUUGAACUAAACCGAGCGCUCGGCCACUGCUCGACAAAGUG-CCAGUCGC------------------ ....(((((..............(((((((.(((((...))))))))))))(((((....)).)))....)))-))......------------------ ( -26.20, z-score = -0.72, R) >droPer1.super_41 721498 81 - 728894 CAAGGGCACGUCCGAUCUUAAAGGCGUUCGCGUUUGAACUAAACCGAGCGCUCGGCCACUGCUCGACAAAGUU-CCAGUCGC------------------ ...(((((.((((((........(((((((.(((((...))))))))))))))))..)))))))(((......-...)))..------------------ ( -24.80, z-score = -0.73, R) >droWil1.scaffold_180949 1042644 93 + 6375548 GAUGGGCACGUCCAAGCCAUGCGACAGUCGUGUUUGAACU-AACCGAGCCAAUGUGCUCGGCAAGGUUGAGUGGUGGAUGGAUCGCUGGAUGGA------ ..(.(((..(((((.(((((.((((..(((....))).((-..((((((......))))))..)))))).)))))...))))).))).).....------ ( -30.70, z-score = -0.28, R) >droVir3.scaffold_13049 9951610 79 + 25233164 --AGAUCACGUCGUCGCUCGGCAACGUUCGCGUUUCAACUAAACCGAGCGUUUGGCCAGAGCUCGACAAGCUGCUUGUUGG------------------- --.(((......)))(((((((.(((((((.((((.....)))))))))))...))).)))).(((((((...))))))).------------------- ( -28.90, z-score = -1.79, R) >droMoj3.scaffold_6680 21580769 73 - 24764193 --UGAUCACGUCGUCGUCCAGCAACGUUCGCGUUUCAACUAAACCGAGCGUCUGCCCAGAGCUCUGCAAGCUGCU------------------------- --.....(((....))).((((.(((((((.((((.....))))))))))).....(((....)))...))))..------------------------- ( -17.70, z-score = 0.01, R) >consensus CAGGGUCACGUCCGAUCCCGGCGACGCUCGCGUUUGAACUAAACCGAGCAUUCGACGACCGCUCGACAAGGUGAUCGACGAUCCACCAA_GUAG______ .........(((.........(((((((((.((((.....))))))))))).))..........)))................................. (-11.32 = -11.05 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:32 2011