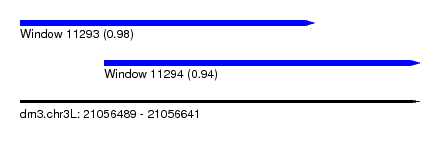
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,056,489 – 21,056,641 |
| Length | 152 |
| Max. P | 0.980108 |

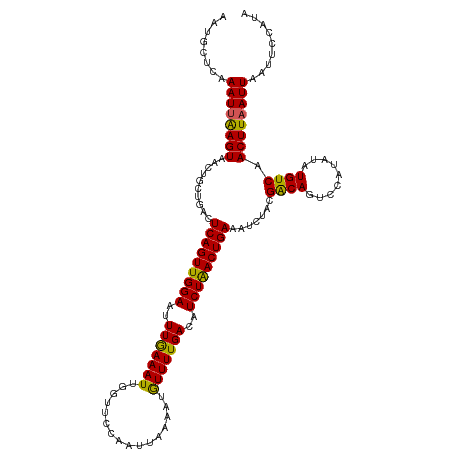
| Location | 21,056,489 – 21,056,601 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Shannon entropy | 0.14338 |
| G+C content | 0.31167 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -22.43 |
| Energy contribution | -21.83 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21056489 112 + 24543557 --------UAGCGUGGUUAUUUAAUGAUUGCGAUCAACGAAAUUCUUAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAA --------....(..((((((((((((((.((.....)).)))).....))))))))))..)....(((((((((......(((((....)))))....(((....)))))))))))).. ( -25.60, z-score = -2.58, R) >droEre2.scaffold_4784 20743995 112 + 25762168 --------UAGCGUGGUUAUUUAAUGAUUGAGUUUAACGAAAUGCUAAAAUGGAGUAGCUGCUGACUCAGUUGGAAUUUGAAAUUGCUUCCAAUUAAAAUGUUUUGACAUCUGACUGAAA --------....(..(((((((.........((((....)))).........)))))))..)....(((((..((..(..((((................))))..)..))..))))).. ( -21.36, z-score = -0.63, R) >droYak2.chr3L 4725952 120 - 24197627 UCUGCAUCUAGCGUGGUUAUUUAAUGAUUGAGUUAAACGAAAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUAAAAUUGCUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAA ...((.....))(..((((((((((..((((((..........))))))))))))))))..)....(((((((((..(((((((................)))))))..))))))))).. ( -30.69, z-score = -3.83, R) >droSec1.super_11 944914 112 + 2888827 --------UAGCGUGGUUAUUUAAUGAUUGCGAUUAACGAAAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUCGUUCCAAUUAAAAUAUUUUGACAUCUAACUGAAA --------....(..((((((((((....(((.((....)).)))....))))))))))..)....(((((((((..(..((((...((......))...))))..)..))))))))).. ( -26.50, z-score = -3.09, R) >droSim1.chr3L 20410525 112 + 22553184 --------UAGCGUGGUUAUUUAAUGAUUGCGAUUAACGAAAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAA --------....(..((((((((((....(((.((....)).)))....))))))))))..)....(((((((((......(((((....)))))....(((....)))))))))))).. ( -28.30, z-score = -3.41, R) >consensus ________UAGCGUGGUUAUUUAAUGAUUGCGAUUAACGAAAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAA ............(..((((((((((........................))))))))))..)....(((((((((..(((((((................)))))))..))))))))).. (-22.43 = -21.83 + -0.60)

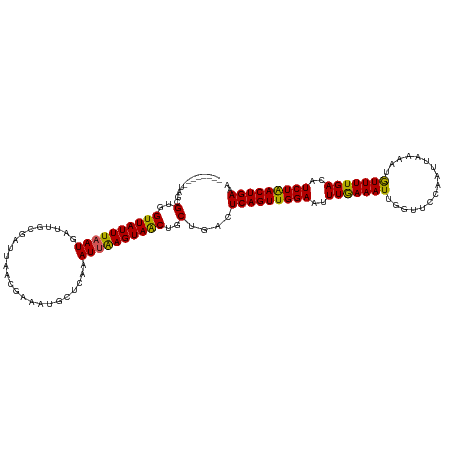
| Location | 21,056,521 – 21,056,641 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.42 |
| Shannon entropy | 0.09793 |
| G+C content | 0.31000 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -21.05 |
| Energy contribution | -20.45 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.938190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21056521 120 + 24543557 AAUUCUUAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAAUCUACGACAGUCCAUAUAUGUCAACUUAAUUAAUUCCAUA ........((((((((..........(((((((((......(((((....)))))....(((....)))))))))))).......((((.........)))).))))))))......... ( -23.20, z-score = -2.32, R) >droEre2.scaffold_4784 20744027 120 + 25762168 AAUGCUAAAAUGGAGUAGCUGCUGACUCAGUUGGAAUUUGAAAUUGCUUCCAAUUAAAAUGUUUUGACAUCUGACUGAAAUCCACGGCAGUCCAUAUAUGUCAACUUAAUUAAUUCCAUA .........(((((((.(((((((..(((((..((..(..((((................))))..)..))..)))))......))))))).((....))............))))))). ( -24.99, z-score = -1.21, R) >droYak2.chr3L 4725992 120 - 24197627 AAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUAAAAUUGCUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAAUCCACGACAGUCCAUAUAUGUCAACUUAAUUAAUUCCAUA ........((((((((..........(((((((((..(((((((................)))))))..))))))))).......((((.........)))).))))))))......... ( -21.89, z-score = -2.45, R) >droSec1.super_11 944946 120 + 2888827 AAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUCGUUCCAAUUAAAAUAUUUUGACAUCUAACUGAAAUCUGCGACAGCCCAUAUAUGUCAACUUAAUUAAUUCCAUA ........((((((((....((.((.(((((((((..(..((((...((......))...))))..)..)))))))))..)).))((((.........)))).))))))))......... ( -24.30, z-score = -3.04, R) >droSim1.chr3L 20410557 120 + 22553184 AAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAAUCUGCGACAGCCCAUAUAUGUCAACUUAAUUAAUUCCAUA ........((((((((....((.((.(((((((((......(((((....)))))....(((....))))))))))))..)).))((((.........)))).))))))))......... ( -26.10, z-score = -2.75, R) >consensus AAUGCUCAAAUUAAGUAACUGCUGACUCAGUUGGAAUUUGAAAUUGGUUCCAAUUAAAAUGUUUUGACAUCUAACUGAAAUCUACGACAGUCCAUAUAUGUCAACUUAAUUAAUUCCAUA ........((((((((..........(((((((((..(((((((................)))))))..))))))))).......((((.........)))).))))))))......... (-21.05 = -20.45 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:29 2011