
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,054,230 – 21,054,368 |
| Length | 138 |
| Max. P | 0.601665 |

| Location | 21,054,230 – 21,054,368 |
|---|---|
| Length | 138 |
| Sequences | 3 |
| Columns | 160 |
| Reading direction | forward |
| Mean pairwise identity | 53.12 |
| Shannon entropy | 0.64115 |
| G+C content | 0.36173 |
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -7.60 |
| Energy contribution | -7.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
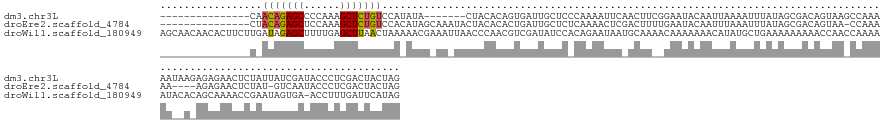
>dm3.chr3L 21054230 138 + 24543557 ---------------CAACAGAGCCCCAAAGCUCUGUCCAUAUA-------CUACACAGUGAUUGCUCCCAAAAUUCAACUUCGGAAUACAAUUAAAAUUUAUAGCGACAGUAAGCCAAAAAUAAGAGAGAACUCUAUUAUCGAUACCCUCGACUACUAG ---------------..(((((((......))))))).......-------........((((((.(((.((........)).)))...))))))..............((((...........((((....))))....((((.....)))).)))).. ( -20.40, z-score = -1.40, R) >droEre2.scaffold_4784 20741667 139 + 25762168 ---------------CUACAGAGCUCCAAAGCUCUGUCCACAUAGCAAAUACUACACACUGAUUGCUCUCAAAACUCGACUUUUGAAUACAAUUUAAAUUUAUAGCGACAGUAA-CCAAAAA----AGAGAACUCUAU-GUCAAUACCCUCGACUACUAG ---------------..((((((((....))))))))..((((((............((((.(((((.((((((......)))))).................)))))))))..-.(.....----.)......))))-))................... ( -20.87, z-score = -1.54, R) >droWil1.scaffold_180949 1034924 159 - 6375548 AGCAACAACACUUCUUGAUAGAGCUUUUGAGCUUAACUAAAAACGAAAUUAACCCAACGUCGAUAUCCACAGAAUAAUGCAAAACAAAAAAACAUAUGCUGAAAAAAAAACCAACCAAAAAUACACAGCAAAACCGAAUAGUGA-ACCUUUGAUUCAUAG .(((.......((((.(((((((((....))))).........(((..((.....))..))).))))...))))...)))................(((((........................)))))..........((((-(.......))))).. ( -14.76, z-score = -0.23, R) >consensus _______________CAACAGAGCUCCAAAGCUCUGUCCAAAUAG_AA_UACUACACAGUGAUUGCUCACAAAAUUCAACUUAAGAAUACAAUUUAAAUUUAUAGCGACAGUAA_CCAAAAA__A_AGAGAACUCUAUUAUCGAUACCCUCGACUACUAG .................(((((((......)))))))........................................................................................................................... ( -7.60 = -7.60 + 0.00)

| Location | 21,054,230 – 21,054,368 |
|---|---|
| Length | 138 |
| Sequences | 3 |
| Columns | 160 |
| Reading direction | reverse |
| Mean pairwise identity | 53.12 |
| Shannon entropy | 0.64115 |
| G+C content | 0.36173 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -13.38 |
| Energy contribution | -12.97 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
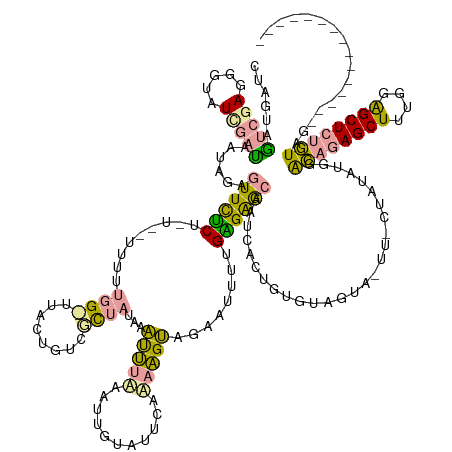
>dm3.chr3L 21054230 138 - 24543557 CUAGUAGUCGAGGGUAUCGAUAAUAGAGUUCUCUCUUAUUUUUGGCUUACUGUCGCUAUAAAUUUUAAUUGUAUUCCGAAGUUGAAUUUUGGGAGCAAUCACUGUGUAG-------UAUAUGGACAGAGCUUUGGGGCUCUGUUG--------------- .(((((((((((((((........((((....)))))))))))))).)))))..((((((.......(((((.(((((((((...)))))))))))))).....)))))-------).....(((((((((....))))))))).--------------- ( -39.90, z-score = -2.56, R) >droEre2.scaffold_4784 20741667 139 - 25762168 CUAGUAGUCGAGGGUAUUGAC-AUAGAGUUCUCU----UUUUUGG-UUACUGUCGCUAUAAAUUUAAAUUGUAUUCAAAAGUCGAGUUUUGAGAGCAAUCAGUGUGUAGUAUUUGCUAUGUGGACAGAGCUUUGGAGCUCUGUAG--------------- ......(((((.....)))))-((((((((((..----.....((-((.(((((.............(((((.(((((((......))))))).)))))....(..((((....))))..).)))))))))..))))))))))..--------------- ( -36.60, z-score = -1.47, R) >droWil1.scaffold_180949 1034924 159 + 6375548 CUAUGAAUCAAAGGU-UCACUAUUCGGUUUUGCUGUGUAUUUUUGGUUGGUUUUUUUUUCAGCAUAUGUUUUUUUGUUUUGCAUUAUUCUGUGGAUAUCGACGUUGGGUUAAUUUCGUUUUUAGUUAAGCUCAAAAGCUCUAUCAAGAAGUGUUGUUGCU ..((.((((((((((-.(((..............))).)))))))))).))........((((((((((...........))))..((((...((((..(...(((((((((((........)))).)))))))...)..)))).))))))))))..... ( -27.74, z-score = 0.23, R) >consensus CUAGUAGUCGAGGGUAUCGAUAAUAGAGUUCUCU_U__UUUUUGG_UUACUGUCGCUAUAAAUUUAAAUUGUAUUCAAAAGUAGAAUUUUGAGAGCAAUCACUGUGUAGUA_UU_CUAUAUGGACAGAGCUUUGGAGCUCUGUAG_______________ (((...(((((.....)))))..))).((((((.........((((........))))...(((((...........)))))........))))))...........................((((((((....))))))))................. (-13.38 = -12.97 + -0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:28 2011