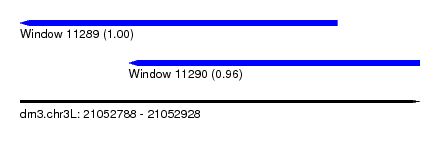
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,052,788 – 21,052,928 |
| Length | 140 |
| Max. P | 0.997785 |

| Location | 21,052,788 – 21,052,899 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.48 |
| Shannon entropy | 0.52127 |
| G+C content | 0.34897 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -14.40 |
| Energy contribution | -15.30 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.997785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
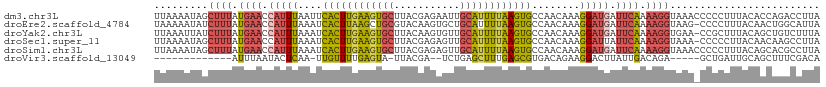
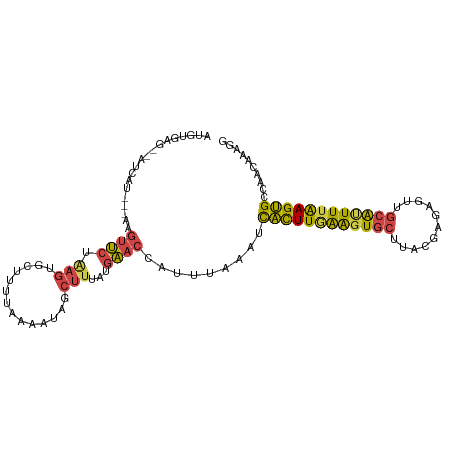
>dm3.chr3L 21052788 111 - 24543557 UUAAAAUAGCUUUAUGAACCAUUUAAUUCACUUGAAGUGCUUACGAGAAUUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUAAACCCCCUUUACACCAGACCUUA ........(((((.((((.(((......((((((((((((...........))))))))))))((......))))).)))).)))))........................ ( -22.40, z-score = -2.74, R) >droEre2.scaffold_4784 20740138 110 - 25762168 UAAAAAUAUCUUUAUGAACCAUUUAAAUCACUUAAGCUGCGUACAAGUGCUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUAAG-CCCCUUUACAACUGGCAUUA ......(((((((.((((.(((......((((((((.((((((....))).))).))))))))((......))))).))))))))))).(-((...........))).... ( -24.60, z-score = -1.90, R) >droYak2.chr3L 4722318 110 + 24197627 UUAAAUUAUCUUUAUGAACCAUUUAAAUCACUUGAAGUGCUUACAAGUGUUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUGAA-CCGCUUUACAGCUGUCUUUA .....((((((((.((((.(((......((((((((((((..((....)).))))))))))))((......))))).))))))))))))(-(.(((....))).))..... ( -27.00, z-score = -2.58, R) >droSec1.super_11 941296 110 - 2888827 UUAAAAUAGCUUUAUGAACCAUUUAAAUCACUUGAAGUGCUUACGAGAGUUGCAUUUUAAGUGCCAACAAAGGAUUAUUCAAAAGGUAAA-CCCCCUUACAACAAGCCUUA ........((((..((((..........((((((((((((..((....)).))))))))))))((......))....)))).((((....-...)))).....)))).... ( -24.30, z-score = -2.97, R) >droSim1.chr3L 20406872 111 - 22553184 UUAAAAUAGCUUUAUGAACCAUUUAAAUCACUUGAAGUGCUUACGAGAGUUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUAAACCCCCUUUACAGCACGCCUUA ........(((...((((.(((......((((((((((((..((....)).))))))))))))((......))))).))))(((((.......)))))..)))........ ( -27.00, z-score = -3.37, R) >droVir3.scaffold_13049 9945572 89 + 25233164 -------------AUUUAAUACUCAA-UUGUUUUGAGUA-UUACGA--UCUGAGCUUUGAGCGUGACAGAAGGACUUAUUGACAGA-----GCUGAUUGCAGCUUUCGACA -------------...((((((((((-.....)))))))-)))(((--((((..(..(((((.(......).).))))..).))))-----((((....))))..)))... ( -23.30, z-score = -1.98, R) >consensus UUAAAAUAGCUUUAUGAACCAUUUAAAUCACUUGAAGUGCUUACGAGAGUUGCAUUUUAAGUGCCAACAAAGGAUGAUUCAAAAGGUAAA_CCCCUUUACAGCAGGCCUUA .........((((.((((.(((((....((((((((((((...........))))))))))))........))))).)))).))))......................... (-14.40 = -15.30 + 0.90)

| Location | 21,052,826 – 21,052,928 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 72.56 |
| Shannon entropy | 0.50945 |
| G+C content | 0.33810 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -12.63 |
| Energy contribution | -12.52 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
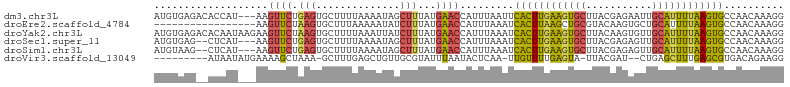
>dm3.chr3L 21052826 102 - 24543557 AUGUGAGACACCAU---AAGUUCUGAGUGCUUUUAAAAUAGCUUUAUGAACCAUUUAAUUCACUUGAAGUGCUUACGAGAAUUGCAUUUUAAGUGCCAACAAAGG .((((......)))---).((((((((.(((........))))))).)))).........((((((((((((...........)))))))))))).......... ( -21.80, z-score = -1.47, R) >droEre2.scaffold_4784 20740175 88 - 25762168 -----------------AAGUUCUAAGUGCUUUAAAAAUAUCUUUAUGAACCAUUUAAAUCACUUAAGCUGCGUACAAGUGCUGCAUUUUAAGUGCCAACAAAGG -----------------..(((.((((((.((((.(((....))).)))).))))))...((((((((.((((((....))).))).))))))))..)))..... ( -16.30, z-score = -0.84, R) >droYak2.chr3L 4722355 105 + 24197627 AUGUGAGACACAAUAAGAAGUUCUAAGUGCUUUUAAAUUAUCUUUAUGAACCAUUUAAAUCACUUGAAGUGCUUACAAGUGUUGCAUUUUAAGUGCCAACAAAGG .((((((.(((....((.....))(((((..(((((((..((.....))...))))))).)))))...)))))))))..((((((((.....))).))))).... ( -22.20, z-score = -1.36, R) >droSec1.super_11 941333 100 - 2888827 AUGUGAG--CUCAU---AAGUUCUGAGUGCUUUUAAAAUAGCUUUAUGAACCAUUUAAAUCACUUGAAGUGCUUACGAGAGUUGCAUUUUAAGUGCCAACAAAGG .(((..(--(....---..((((((((.(((........))))))).))))..........(((((((((((..((....)).)))))))))))))..))).... ( -25.90, z-score = -2.20, R) >droSim1.chr3L 20406910 100 - 22553184 AUGUAAG--CUCAU---AAGUUCUGAGUGCUUUUAAAAUAGCUUUAUGAACCAUUUAAAUCACUUGAAGUGCUUACGAGAGUUGCAUUUUAAGUGCCAACAAAGG .(((..(--(....---..((((((((.(((........))))))).))))..........(((((((((((..((....)).)))))))))))))..))).... ( -24.90, z-score = -2.10, R) >droVir3.scaffold_13049 9945605 91 + 25233164 ---------AUAAUAUGAAAAGCUAAA-GCUUUGAGCUGUUGCGUAUUUAAUACUCAA-UUGUUUUGAGUA-UUACGAU--CUGAGCUUUGAGCGUGACAGAAGG ---------.........(((((....-)))))...((((..(((...((((((((((-.....)))))))-)))(((.--.......))).)))..)))).... ( -26.10, z-score = -2.97, R) >consensus AUGUGAG__AUCAU___AAGUUCUAAGUGCUUUUAAAAUAGCUUUAUGAACCAUUUAAAUCACUUGAAGUGCUUACGAGAGUUGCAUUUUAAGUGCCAACAAAGG ...................((((.(((..............)))...)))).........((((((((((((...........)))))))))))).......... (-12.63 = -12.52 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:26 2011