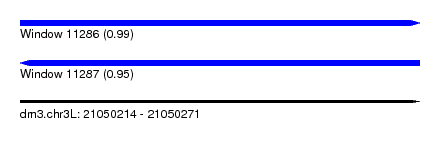
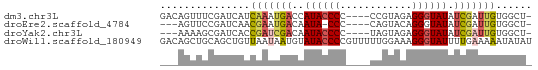
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,050,214 – 21,050,271 |
| Length | 57 |
| Max. P | 0.986324 |

| Location | 21,050,214 – 21,050,271 |
|---|---|
| Length | 57 |
| Sequences | 4 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 59.60 |
| Shannon entropy | 0.63251 |
| G+C content | 0.43205 |
| Mean single sequence MFE | -16.12 |
| Consensus MFE | -6.35 |
| Energy contribution | -6.73 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
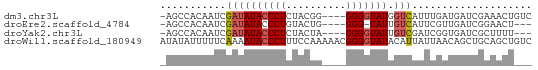
>dm3.chr3L 21050214 57 + 24543557 -AGCCACAAUCGAUAUACCCUCUACGG----GGGGUAUGGUCAUUUGAUGAUCGAAACUGUC -........(((((((((((((....)----))))))).(((....))).)))))....... ( -19.40, z-score = -2.45, R) >droEre2.scaffold_4784 20737502 53 + 25762168 -AGCCACAAUCGAUAUACCCUGUACUG----GGG-UAUUGUCAUUCGUUGAUCGGAACU--- -..........((((((((((.....)----)))-)).)))).((((.....))))...--- ( -13.10, z-score = -0.82, R) >droYak2.chr3L 4719804 54 - 24197627 -AGCCACAAUCGAUAUACCCUCUACUA----GGGGUAUUGUCGAUCGGUGAUCGCUUUU--- -((((((.(((((((((((((......----)))))).)))))))..)))...)))...--- ( -18.80, z-score = -3.19, R) >droWil1.scaffold_180949 1031521 62 - 6375548 AUAUAUUUUUCAAAAUACCCUUUCCAAAAACGGGGUAUACAUUAUUAACAGCUGCAGCUGUC ..............(((((((..........))))))).........(((((....))))). ( -13.20, z-score = -2.82, R) >consensus _AGCCACAAUCGAUAUACCCUCUACUA____GGGGUAUUGUCAAUCGAUGAUCGCAACU___ ...........((((((((((..........))))))).))).................... ( -6.35 = -6.73 + 0.38)
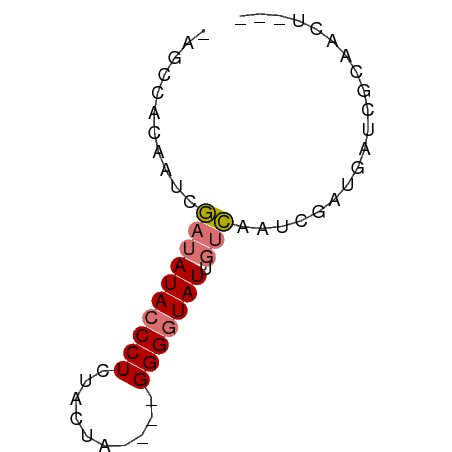
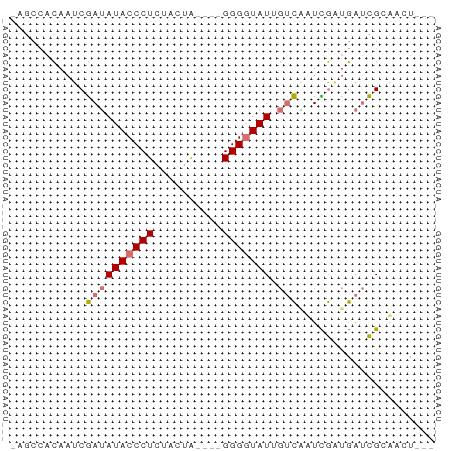
| Location | 21,050,214 – 21,050,271 |
|---|---|
| Length | 57 |
| Sequences | 4 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 59.60 |
| Shannon entropy | 0.63251 |
| G+C content | 0.43205 |
| Mean single sequence MFE | -14.40 |
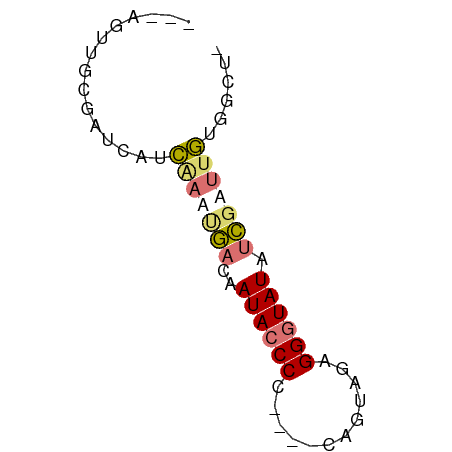
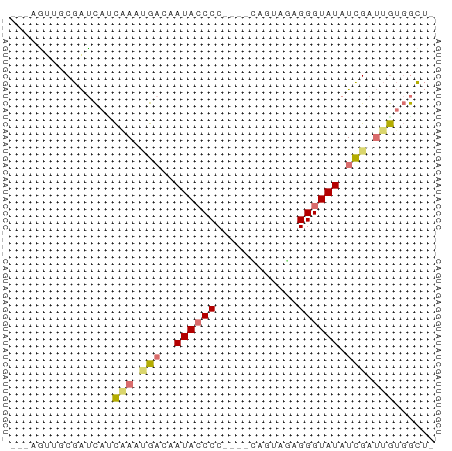
| Consensus MFE | -6.28 |
| Energy contribution | -7.03 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21050214 57 - 24543557 GACAGUUUCGAUCAUCAAAUGACCAUACCCC----CCGUAGAGGGUAUAUCGAUUGUGGCU- ...(((..(((((.((....))..((((((.----.......))))))...)))))..)))- ( -13.90, z-score = -1.07, R) >droEre2.scaffold_4784 20737502 53 - 25762168 ---AGUUCCGAUCAACGAAUGACAAUA-CCC----CAGUACAGGGUAUAUCGAUUGUGGCU- ---(((..(((((...(.....).(((-(((----.......))))))...)))))..)))- ( -12.90, z-score = -1.18, R) >droYak2.chr3L 4719804 54 + 24197627 ---AAAAGCGAUCACCGAUCGACAAUACCCC----UAGUAGAGGGUAUAUCGAUUGUGGCU- ---...(((...(((.((((((..(((((((----.....).)))))).))))))))))))- ( -16.80, z-score = -2.76, R) >droWil1.scaffold_180949 1031521 62 + 6375548 GACAGCUGCAGCUGUUAAUAAUGUAUACCCCGUUUUUGGAAAGGGUAUUUUGAAAAAUAUAU ((((((....))))))........((((((..((....))..)))))).............. ( -14.00, z-score = -1.84, R) >consensus ___AGUUGCGAUCAUCAAAUGACAAUACCCC____CAGUAGAGGGUAUAUCGAUUGUGGCU_ ...............(((((((..((((((............)))))).)))))))...... ( -6.28 = -7.03 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:23 2011