
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,036,067 – 21,036,182 |
| Length | 115 |
| Max. P | 0.812430 |

| Location | 21,036,067 – 21,036,169 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.31 |
| Shannon entropy | 0.09862 |
| G+C content | 0.45957 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
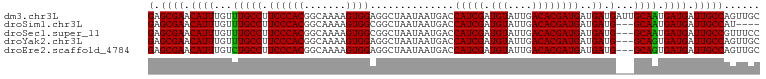
>dm3.chr3L 21036067 102 - 24543557 GAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGAGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUGAUUGCAAUGAUGAUUGCCAGUUGC ..((((.((((((((.((((..((((.......)))))))).)))).....(((((.(((....))))))))..))))...(((((....)))))...)))) ( -26.10, z-score = -1.32, R) >droSim1.chr3L 20390013 95 - 22553184 GAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGCGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG---GCAAUGAUGAUUGCCAU---- .......((((.(((.(((...((((.......)))).))).)))))))..(((((.(((....))))))))...(((---(((((....))))))))---- ( -28.10, z-score = -2.03, R) >droSec1.super_11 924452 99 - 2888827 GAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGCGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG---GCAAUGAUGAUUGCCGUUUCC .......((((.(((.(((...((((.......)))).))).)))))))..(((((.(((....))))))))..((((---(((((....)))))))))... ( -28.90, z-score = -1.81, R) >droYak2.chr3L 4705150 99 + 24197627 GAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGAGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG---GCAGUGAUGAUUGCCAGUUGC ..((((.((((.(((.((((..((((.......)))))))).)))))))..(((((.(((....))))))))....((---(((((....))))))).)))) ( -31.00, z-score = -2.59, R) >droEre2.scaffold_4784 20717741 99 - 25762168 GAGCGAACAUUUGUCUGCCUUCCCACGGCAAAAGUGGAGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG---GCAGUGAUGAUUGCCAGUUGC ..((((.((((.((..((((..((((.......))))))))..))))))..(((((.(((....))))))))....((---(((((....))))))).)))) ( -30.50, z-score = -2.12, R) >consensus GAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGAGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG___GCAAUGAUGAUUGCCAGUUGC ..((((((....))))))........(((((..((.(..((.....((.(.(((((.(((....)))))))).).))....))..).))..)))))...... (-24.26 = -24.46 + 0.20)


| Location | 21,036,090 – 21,036,182 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 92.16 |
| Shannon entropy | 0.12826 |
| G+C content | 0.45006 |
| Mean single sequence MFE | -18.52 |
| Consensus MFE | -17.34 |
| Energy contribution | -17.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
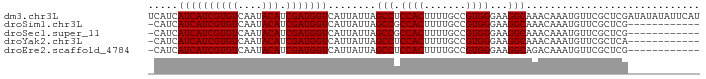
>dm3.chr3L 21036090 92 + 24543557 UCAUCAUCAUCGUGUCAAUACAUCGAUGGUCAUUAUUAGCCUCCACUUUUGCCGUGGGAAGGCAAACAAAUGUUCGCUCGAUAUAUAUUCAU ..(((((((((((((....))).)))))))........((((((((.......))))..))))................))).......... ( -19.80, z-score = -1.32, R) >droSim1.chr3L 20390030 79 + 22553184 -CAUCAUCAUCGUGUCAAUACAUCGAUGGUCAUUAUUAGCCGCCACUUUUGCCGUGGGAAGGCAAACAAAUGUUCGCUCG------------ -....((((((((((....))).)))))))........(((.((((.......))))...))).................------------ ( -18.00, z-score = -0.83, R) >droSec1.super_11 924473 79 + 2888827 -CAUCAUCAUCGUGUCAAUACAUCGAUGGUCAUUAUUAGCCGCCACUUUUGCCGUGGGAAGGCAAACAAAUGUUCGCUCG------------ -....((((((((((....))).)))))))........(((.((((.......))))...))).................------------ ( -18.00, z-score = -0.83, R) >droYak2.chr3L 4705171 79 - 24197627 -CAUCAUCAUCGUGUCAAUACAUCGAUGGUCAUUAUUAGCCUCCACUUUUGCCGUGGGAAGGCAAACAAAUGUUCGCUCA------------ -....((((((((((....))).)))))))........((((((((.......))))..)))).................------------ ( -18.40, z-score = -1.47, R) >droEre2.scaffold_4784 20717762 79 + 25762168 -CAUCAUCAUCGUGUCAAUACAUCGAUGGUCAUUAUUAGCCUCCACUUUUGCCGUGGGAAGGCAGACAAAUGUUCGCUCG------------ -....((((((((((....))).)))))))........((((((((.......))))..)))).................------------ ( -18.40, z-score = -1.00, R) >consensus _CAUCAUCAUCGUGUCAAUACAUCGAUGGUCAUUAUUAGCCUCCACUUUUGCCGUGGGAAGGCAAACAAAUGUUCGCUCG____________ .....((((((((((....))).)))))))........(((.((((.......))))...)))............................. (-17.34 = -17.34 + -0.00)
| Location | 21,036,090 – 21,036,182 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Shannon entropy | 0.12826 |
| G+C content | 0.45006 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
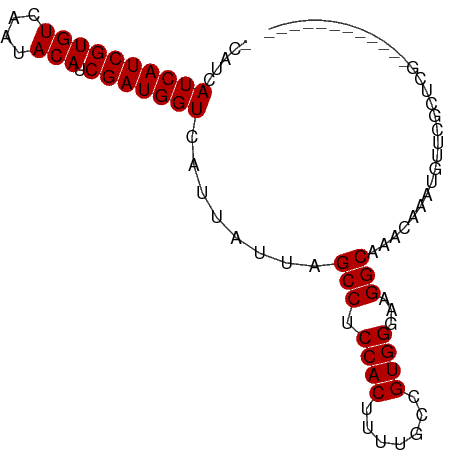
>dm3.chr3L 21036090 92 - 24543557 AUGAAUAUAUAUCGAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGAGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUGA .........(((((......((((.(((.((((..((((.......)))))))).)))))))..(((((.(((....)))))))).))))). ( -23.10, z-score = -1.90, R) >droSim1.chr3L 20390030 79 - 22553184 ------------CGAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGCGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG- ------------........((((.(((.(((...((((.......)))).))).)))))))..(((((.(((....))))))))......- ( -19.10, z-score = -0.94, R) >droSec1.super_11 924473 79 - 2888827 ------------CGAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGCGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG- ------------........((((.(((.(((...((((.......)))).))).)))))))..(((((.(((....))))))))......- ( -19.10, z-score = -0.94, R) >droYak2.chr3L 4705171 79 + 24197627 ------------UGAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGAGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG- ------------........((((.(((.((((..((((.......)))))))).)))))))..(((((.(((....))))))))......- ( -20.60, z-score = -2.05, R) >droEre2.scaffold_4784 20717762 79 - 25762168 ------------CGAGCGAACAUUUGUCUGCCUUCCCACGGCAAAAGUGGAGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG- ------------........((((.((..((((..((((.......))))))))..))))))..(((((.(((....))))))))......- ( -20.10, z-score = -1.53, R) >consensus ____________CGAGCGAACAUUUGUUUGCCUUCCCACGGCAAAAGUGGAGGCUAAUAAUGACCAUCGAUGUAUUGACACGAUGAUGAUG_ ....................((((.(((.((((..((((.......)))))))).)))))))..(((((.(((....))))))))....... (-18.98 = -19.58 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:20 2011