
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,028,424 – 21,028,539 |
| Length | 115 |
| Max. P | 0.689786 |

| Location | 21,028,424 – 21,028,539 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 61.59 |
| Shannon entropy | 0.67860 |
| G+C content | 0.32420 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -8.34 |
| Energy contribution | -10.06 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.597075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
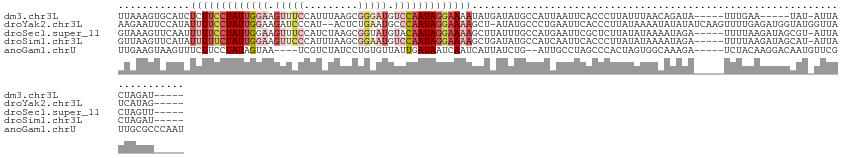
>dm3.chr3L 21028424 115 + 24543557 -----AUCUAGUAAU-AUA-----UUCAAA-----UAUCUGUUAAAUAAGGGUGAAUUAAUGGCAUAUCAUAUUUUCCUAUUGGACAUCCCGCUUAAAUGGAAACUUCCAAUAGGAAGAGAUGCACUUUAA -----.....((...-..(-----((((..-----..(((........))).))))).....))....(((.(((((((((((((....(((......))).....))))))))))))).)))........ ( -25.20, z-score = -1.68, R) >droYak2.chr3L 4697302 123 - 24197627 -----CUAUGAUAACCAUACCAUCUCAAAACUUGAUAUAUAUUUUAUAAGGGUGAAUUCAGGGCAUAU-AGCUUUUCCUAUUGGGCAUUCAGAGU--AUGGGAUCUUCCAAUAGGAAAAUAUGGAAUUCUU -----.((((.....))))..........((((..((((.....)))).))))((((((..(((....-.)))((((((((((((.((((.....--...))))..)))))))))))).....)))))).. ( -26.20, z-score = -0.31, R) >droSec1.super_11 916739 120 + 2888827 -----AACUAGUAAU-ACGCUAUCUUAAAA-----UCUAUUUUAUAUAAGAGCGAAUUCAUGGCAAAUAAGCUUUUCCUAUUGUACAUACCGCUUAGAUGGAAACUUCCAAUAGGAAAAAUUGAACUUUAC -----.....((((.-.((((....(((((-----....)))))......))))..((((..((......))(((((((((((......(((......))).......)))))))))))..))))..)))) ( -19.92, z-score = -1.18, R) >droSim1.chr3L 20374953 120 + 22553184 -----AUCUAGUAAU-AUGCUAUCUUAAAA-----UCUAUUUUAUAUAAGGGUGAAUUGAUGGCAUAUCAGCUUUUCCUAUUGGACAUUCCGCUUAAAUGGGAACUUCCAAUAGAAAAAUAUGAACUUAAC -----.........(-((((((((.....(-----(((...........)))).....)))))))))(((..((((.((((((((..((((.(......)))))..)))))))).))))..)))....... ( -30.60, z-score = -3.67, R) >anoGam1.chrU 54890818 120 + 59568033 AUUGGGCGCAACGAACAUUGUCCUUGUAGA-----UCUUUGCCACUAGUGGGCUAGGCAAU--CAGAUAAUGAUUGAUUAUCAAUAACACAGGAUAGACGA----UUACUAUAGGAAGAAACUUACUUCAA ...(((((..........)))))(((((((-----((((((((..(((....)))))))).--.))))..((((((.(((((..........))))).)))----)))))))))((((.......)))).. ( -24.10, z-score = -0.46, R) >consensus _____AUCUAGUAAU_AUACUAUCUUAAAA_____UCUAUGUUAUAUAAGGGUGAAUUAAUGGCAUAUAAGCUUUUCCUAUUGGACAUACCGCUUAAAUGGAAACUUCCAAUAGGAAAAAAUGAACUUCAA ........................................................................(((((((((((((....(((......))).....)))))))))))))............ ( -8.34 = -10.06 + 1.72)


| Location | 21,028,424 – 21,028,539 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 61.59 |
| Shannon entropy | 0.67860 |
| G+C content | 0.32420 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -8.24 |
| Energy contribution | -11.52 |
| Covariance contribution | 3.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
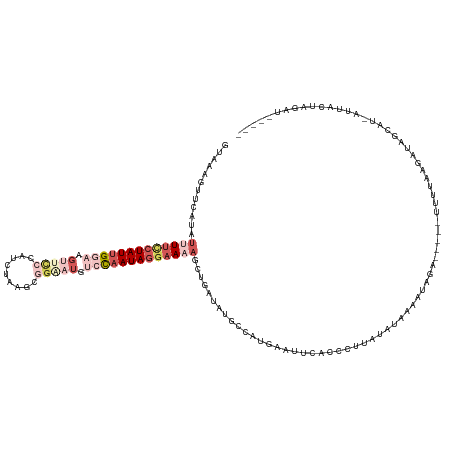
>dm3.chr3L 21028424 115 - 24543557 UUAAAGUGCAUCUCUUCCUAUUGGAAGUUUCCAUUUAAGCGGGAUGUCCAAUAGGAAAAUAUGAUAUGCCAUUAAUUCACCCUUAUUUAACAGAUA-----UUUGAA-----UAU-AUUACUAGAU----- .....(.((((.(((((((((((((...((((........))))..))))))))))).....)).)))))....(((((....((((.....))))-----..))))-----)..-..........----- ( -26.10, z-score = -2.29, R) >droYak2.chr3L 4697302 123 + 24197627 AAGAAUUCCAUAUUUUCCUAUUGGAAGAUCCCAU--ACUCUGAAUGCCCAAUAGGAAAAGCU-AUAUGCCCUGAAUUCACCCUUAUAAAAUAUAUAUCAAGUUUUGAGAUGGUAUGGUUAUCAUAG----- ..((((((....((((((((((((.(((......--..)))......))))))))))))((.-....))...))))))(((.((.((((((.........)))))).)).))).............----- ( -25.00, z-score = -1.14, R) >droSec1.super_11 916739 120 - 2888827 GUAAAGUUCAAUUUUUCCUAUUGGAAGUUUCCAUCUAAGCGGUAUGUACAAUAGGAAAAGCUUAUUUGCCAUGAAUUCGCUCUUAUAUAAAAUAGA-----UUUUAAGAUAGCGU-AUUACUAGUU----- ((((((((((..(((((((((((...((..((........)).))...)))))))))))((......))..))))))((((((((...........-----...)))))..))).-.)))).....----- ( -24.54, z-score = -1.57, R) >droSim1.chr3L 20374953 120 - 22553184 GUUAAGUUCAUAUUUUUCUAUUGGAAGUUCCCAUUUAAGCGGAAUGUCCAAUAGGAAAAGCUGAUAUGCCAUCAAUUCACCCUUAUAUAAAAUAGA-----UUUUAAGAUAGCAU-AUUACUAGAU----- ............(((((((((((((.((((((......).))))).)))))))))))))..((((((((.(((.......................-----......))).))))-))))......----- ( -28.85, z-score = -3.44, R) >anoGam1.chrU 54890818 120 - 59568033 UUGAAGUAAGUUUCUUCCUAUAGUAA----UCGUCUAUCCUGUGUUAUUGAUAAUCAAUCAUUAUCUG--AUUGCCUAGCCCACUAGUGGCAAAGA-----UCUACAAGGACAAUGUUCGUUGCGCCCAAU ..((((.......)))).....((((----((((...((((.(((....(((...((((((.....))--))))....((((....).)))....)-----)).)))))))..)))...)))))....... ( -18.80, z-score = 0.64, R) >consensus GUAAAGUUCAUAUUUUCCUAUUGGAAGUUCCCAUCUAAGCGGAAUGUCCAAUAGGAAAAGCUGAUAUGCCAUGAAUUCACCCUUAUAUAAAAUAGA_____UUUUAAGAUAGCAU_AUUACUAGAU_____ ............(((((((((((((.(((((.........))))).)))))))))))))........................................................................ ( -8.24 = -11.52 + 3.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:18 2011