| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,022,571 – 21,022,691 |
| Length | 120 |
| Max. P | 0.614025 |

| Location | 21,022,571 – 21,022,691 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.11 |
| Shannon entropy | 0.53048 |
| G+C content | 0.57667 |
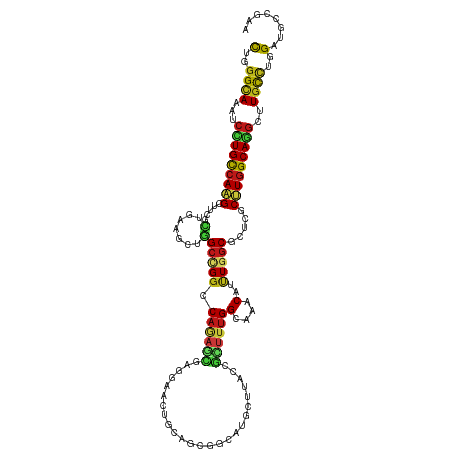
| Mean single sequence MFE | -50.84 |
| Consensus MFE | -21.62 |
| Energy contribution | -20.96 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614025 |
| Prediction | RNA |
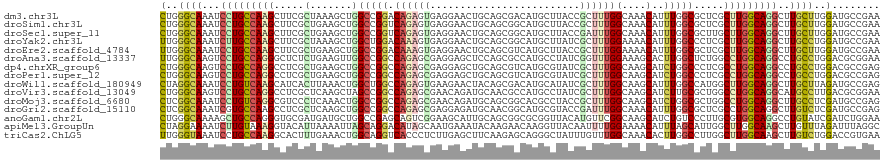
Download alignment: ClustalW | MAF
>dm3.chr3L 21022571 120 + 24543557 CUGGGCAAAUCCUGCCAAGCUUCGCUAAAGCUGGCCGGACAGAGUGAGGAACUGCAGCGACAUGCUUACCGCUUUGGCAAACAUUUGGCGCUCGCUUGGCAGGCUUGCUUGGAUGCCGAA (..(((((..((((((((((..(((((((....((((....(((((.(.....(((......)))...)))))))))).....)))))))...)))))))))).)))))..)........ ( -49.80, z-score = -1.94, R) >droSim1.chr3L 20369107 120 + 22553184 CUGGGCAAAUCCUGCCAAGCUUCGCUGAAGCUGGCCGGUCAGAGUGAGGAACUGCAGCGGCAUGCUUACCGCUUUGGCAAACAUUUGGCGCUCGCUUGGCAGGCUUGCUUGGAUGCCGAA (..(((((..((((((((((..(((..((..((....(((((((((.(.....(((......)))...))))))))))...))))..)))...)))))))))).)))))..)........ ( -51.70, z-score = -1.28, R) >droSec1.super_11 910912 120 + 2888827 CUGGGCAAAUCCUGCCAAGCUUCGCUGAAGCUGGCCGGUCAGAGUGAGGAACUGCAGCGGCAUGCUUACCGAUUUGGCAAACAUUUGGCGCUUGCUUGGCAGGCUUGCUUGGAUGCCGAA (..(((((..((((((((((..(((..((....(((((...(.(((((.(..(((....)))).))))))...))))).....))..)))...)))))))))).)))))..)........ ( -52.30, z-score = -2.03, R) >droYak2.chr3L 4689655 120 - 24197627 UUGGGCAAAUCUUGCCAAGCUUCGCUAAAGCUGGCUGGACAAAGUGAGGAACUGCAGCGGCAUGCUUAUCGCUUUGGAAAACAUUUGGCCCUCGCUUGGCAGGCUUGCCUGGAUGCCGAA (..(((((..((((((((((...((....)).(((..((((((((((......(((......)))...)))))))).......))..)))...)))))))))).)))))..)........ ( -46.31, z-score = -1.28, R) >droEre2.scaffold_4784 20703964 120 + 25762168 UUGGGCAAAUCCUGCCAAGCUUCGCUGAAGCUGGCCGGACAAAGUGAGGAACUGCAGCGUCAUGCUUACCGCUUUGGAAAACAUUUGGCGCUCGCUUGGCAGGCUUGCUUGGAUGCCGAA (..(((((..((((((((((..(((..((..((......(((((((.(.....(((......)))...)))))))).....))))..)))...)))))))))).)))))..)........ ( -45.80, z-score = -0.95, R) >droAna3.scaffold_13337 14619793 120 - 23293914 UUGGGCAAGUCCUGCCAGGGCUCUCUGAAGUUGGCCGGCCAGAGCGAGGAGCUCCAGCGCCAUGCCUAUCGGUUUGGAAAGCACUUGGCUCUGGCCUGGCAGGCCUGCCUGGACGCGGAA (..((((.(.(((((((.((((..(....)..))))(((((((((.(((.(((......(((.(((....))).)))..))).))).))))))))))))))))).))))..)........ ( -62.80, z-score = -2.25, R) >dp4.chrXR_group6 12189102 120 - 13314419 CUGGGCAAGUCCUGCCAGGCCUCGCUGAAGCUGGCCGGCCAGAGCGAGGAGCUGCAGCGUCAUGCGUAUCGCUUUGGCAAGCAUCUGGCCCUCGCCUGGCAGGCCUGCCUGGACGCCGAG (..((((.(.((((((((((...(((((.(((.....((((((((((...((.(((......))))).)))))))))).))).)).)))....))))))))))).))))..)........ ( -62.70, z-score = -1.51, R) >droPer1.super_12 600603 120 - 2414086 CUGGGCAAGUCCUGCCAGGCCUCGCUGAAGCUGGCCGGCCAGAGCGAGGAGCUGCAGCGUCAUGCGUAUCGCUUUGGCAAGCAUCUGGCCCUCGCCUGGCAGGCCUGCCUGGACGCCGAG (..((((.(.((((((((((...(((((.(((.....((((((((((...((.(((......))))).)))))))))).))).)).)))....))))))))))).))))..)........ ( -62.70, z-score = -1.51, R) >droWil1.scaffold_180949 994898 120 - 6375548 CUAGGCAAAUCCUGUCAAGCAUCACUUAAACUGGCUGGCCAGAGUGAAGAACUACAGCGACAUGCAUAUCGCUUUGGCAAGCAUUUGGCCAUGGCUUGGCAGGCUUGCUUAGAUGCCGAG ((((((((..((((((((((.....(((((...(((.((((((((((.(.....).((.....))...)))))))))).))).))))).....)))))))))).))))))))........ ( -47.20, z-score = -2.78, R) >droVir3.scaffold_13049 24530637 120 - 25233164 CUGGGCAAGUCCUGCCAGGCCUCGCUCAAGCUAGCCGGCCAGAGCGAACAGAUGCAACGCCAUGCCUAUCGCUUUGGCAAGCAUCUUGCGCUGGCCUGGCAGGCAUGCCUUGACGCGGAA (.(((((.(.(((((((((((..((.((((...((..((((((((((..((..(((......))))).))))))))))..))..)))).)).)))))))))))).))))).)........ ( -62.50, z-score = -3.53, R) >droMoj3.scaffold_6680 531888 120 + 24764193 CUCGGCAAAUCCUGUCAGGCGUCCCUCAAACUGGCCGGCCAGAGCGAACAGAUGCAGCGGCACGCCUACCGCUUUGGCAAGCAUCUGGCGCUGGCCUGGCAGGCUUGCCUCGAUGCCGAG (((((((((.((((((((((..........((((....))))((((..((((((((((((........))))).......))))))).)))).)))))))))).)).......))))))) ( -56.92, z-score = -2.25, R) >droGri2.scaffold_15110 22628345 120 - 24565398 CUCGGCAAAUCGUGCCAAGCCUCGCUCAAGCUGGCCGGCCAGAGCGAGGAGAUGCAACGGCAUGCGUACCGAUUUGGCAAACAUUUGGCGCUCGCCUGGCAGGCUUGUCUCGAUGCCGAG (((((((..(((.(.((((((((((((..(((....)))..)))))).....(((...(((..(((..((((..((.....)).)))))))..)))..))))))))).).)))))))))) ( -54.20, z-score = -2.15, R) >anoGam1.chr2L 24691972 120 - 48795086 CUGGGCAAAAGCUGCCAGGGUGCGAUGAUGCUGGCCGAGCAGUCGGAAGCAUUGCAGCGGCGCGGUUACAUGUUCGGCAAGCAUCUGUCCCUUGCGUGGCAGGCCUGUAUCGAUCUGGAA ..((((....((..((((((..((..((((((.(((((((((.(....))(((((......)))))....)))))))).))))))))..))))).)..))..)))).............. ( -49.20, z-score = -0.41, R) >apiMel3.GroupUn 38884188 120 + 399230636 CUAGGAAAAUCUUGUAAAGGUACAUUAAAAUUAGCAGGACAUAGCAAUGAAAUACAAGAACAAGGUUACAAUUUUGGAAAACAUUUAGCAUUGGCUUGGCAAGCUUGUUUAGAUUUAGGC (((((..((((((((....(((.......(((.((........)))))....)))....)))))))).((((.(((((.....))))).)))).)))))...(((((........))))) ( -16.40, z-score = 1.16, R) >triCas2.ChLG5 18761456 120 + 18847211 UUGGGUAAAUCCUGCCAAGGCACUUUGAAACUGGCAGGUCACCCUCUUGAGCUUCAAGAGCAGGGCUAUUUGUUUGGCAAACACUUGGCCUUGGCUUGGCAAGCUUGUCUGGACCGUGAA ..((((....(((((((.(...........))))))))..))))(((.((((((((((..((((((((..((((.....))))..)))))))).)))))..)))))....)))....... ( -42.00, z-score = -0.62, R) >consensus CUGGGCAAAUCCUGCCAAGCUUCGCUGAAGCUGGCCGGCCAGAGCGAGGAACUGCAGCGGCAUGCUUACCGCUUUGGCAAACAUUUGGCGCUCGCUUGGCAGGCUUGCCUGGAUGCCGAA (..((((...(((((((((.....(.......)(((((.((((((.........................))))))(....)..))))).....)))))))))..))))..)........ (-21.62 = -20.96 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:16 2011