| Sequence ID | dm3.chr3L |
|---|---|
| Location | 21,001,393 – 21,001,491 |
| Length | 98 |
| Max. P | 0.656028 |

| Location | 21,001,393 – 21,001,491 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.78 |
| Shannon entropy | 0.12732 |
| G+C content | 0.42561 |
| Mean single sequence MFE | -17.90 |
| Consensus MFE | -14.95 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 21001393 98 + 24543557 AAACACACA-CACACAUAUUGACACACUACCCAACGGCGAAAAGUUCAAAGCUCGCCAAAAUUAUAAAUAAAUGUCCGCAAGGGUAGGCCAGGCAAAAC .........-........(((.....((((((...(((((............)))))...............((....)).))))))......)))... ( -18.80, z-score = -1.85, R) >droEre2.scaffold_4784 20690691 95 + 25762168 ---UACACA-CACACACAUUGACACACUACACAACGGCGAAAAGUUCAAAGCUCGCCAAAAUUAUAAAUAAAUGUCCACAAGGGUAGGCCAGGCGAAAC ---......-........(((.....((((.....(((((............)))))..................((....))))))......)))... ( -13.00, z-score = -0.32, R) >droYak2.chr3L 4676082 96 - 24197627 ---CAUAAAACACACACAUUGACACACUACUCAACGGCGAAAAGUUCAAAGCUCGCCAAAAUUAUAAAUAAAUGUCCGCAAGGGUAGGCCUGUCGAAAC ---...............((((((..((((.....(((((............)))))..................((....))))))...))))))... ( -21.90, z-score = -3.08, R) >consensus ___CACACA_CACACACAUUGACACACUACACAACGGCGAAAAGUUCAAAGCUCGCCAAAAUUAUAAAUAAAUGUCCGCAAGGGUAGGCCAGGCGAAAC ..................(((.....((((.....(((((............)))))..................((....))))))......)))... (-14.95 = -14.73 + -0.22)

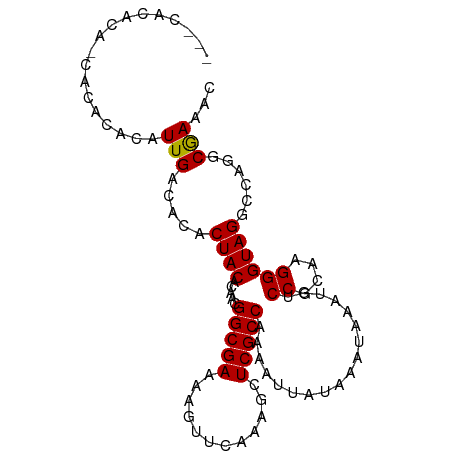

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:14 2011