
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 611,306 – 611,392 |
| Length | 86 |
| Max. P | 0.872291 |

| Location | 611,306 – 611,392 |
|---|---|
| Length | 86 |
| Sequences | 9 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 70.19 |
| Shannon entropy | 0.63465 |
| G+C content | 0.44885 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -8.42 |
| Energy contribution | -8.14 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
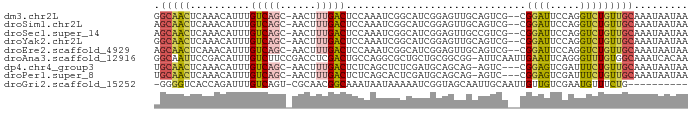
>dm3.chr2L 611306 86 + 23011544 GGCAACUCAAACAUUUGUCAGC-AACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCG--CGGAUUCCAGGUCUGUUGCAAAUAAUAA (((((.........))))).((-((((((((..((.....))..)))))))))).((.(--(((((.....)))))).))......... ( -25.70, z-score = -2.68, R) >droSim1.chr2L 617648 86 + 22036055 AGCAACUCAAACAUUUGUCAGC-AACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCG--CGGAUUCCAGGUCUGUUGCAAAUAAUAA .(((((..............((-((((((((..((.....))..)))))))))).....--.((((.....)))))))))......... ( -24.20, z-score = -2.34, R) >droSec1.super_14 594004 86 + 2068291 AGCAACUCAAACAUUUGUCAGC-AACUUUGACUCCAAAUCGGCAUCGGAGUUGCCGUCG--CGGAUUCCAGGUCUGUUGCAAAUAAUAA ............(((((.((((-(((((.((.(((....(((((.......)))))...--.))).)).)))).))))))))))..... ( -24.50, z-score = -2.26, R) >droYak2.chr2L 600278 86 + 22324452 GGCAACUCAAACAUUUGUCAGC-AACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCG--CGGAUUCCAGGUCUGUUGCAAAUAAUAA (((((.........))))).((-((((((((..((.....))..)))))))))).((.(--(((((.....)))))).))......... ( -25.70, z-score = -2.68, R) >droEre2.scaffold_4929 667359 86 + 26641161 AGCAACUCAAACAUUUGUCAGC-AACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCG--CGGAUUCCAGGUCUGUUGCAAAUAAUAA .(((((..............((-((((((((..((.....))..)))))))))).....--.((((.....)))))))))......... ( -24.20, z-score = -2.34, R) >droAna3.scaffold_12916 1611427 88 + 16180835 GGCAAUUCCGACAUUUGUCUUCCGACCUCGACUGCCAGGCGCUGCUGCGGCGG-AUUCAAUUGAAUUCAGGGUUUGUGGCAAAUCACAA ((((.....(((....)))...((....))..)))).(.(((....))).)((-((((....)))))).....((((((....)))))) ( -23.50, z-score = 0.37, R) >dp4.chr4_group3 9118523 84 + 11692001 UGCAACUCAAACAUUUGUCAGC-AACUUUGACUCUCAGCUCUCGAUGCAGCAG-AGUC---CGGAGUCGAUUUCUGUUGCAAAUAAUAA ............(((((.((((-(...((((((((..((((((......).))-))).---.))))))))....))))))))))..... ( -25.10, z-score = -2.59, R) >droPer1.super_8 302459 84 + 3966273 UGCAACUCAAACAUUUGUCAGC-AACUUUGACUCUCAGCACUCGAUGCAGCAG-AGUC---CGGAGUCGAUUUCUGUUGCAAAUAAUAA ............(((((.((((-(...((((((((..(.((((.........)-))).---)))))))))....))))))))))..... ( -21.00, z-score = -1.22, R) >droGri2.scaffold_15252 6973440 77 + 17193109 -GGGGUCACCAGAUUUGUCAGU-CGCAACGGCAAAUAAUAAAAAUCGGUAGCAAUUGCAAUUGUUGUCGAAUGUUUCUG---------- -.((....))..(((((((.((-....)))))))))........(((..((((((....))))))..))).........---------- ( -16.30, z-score = -0.81, R) >consensus AGCAACUCAAACAUUUGUCAGC_AACUUUGACUCCAAAUCGGCAUCGGAGUUGCAGUCG__CGGAUUCCAGGUCUGUUGCAAAUAAUAA .(((((..........(((((......)))))..............................((((.....)))))))))......... ( -8.42 = -8.14 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:20 2011