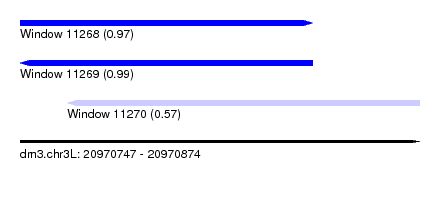
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,970,747 – 20,970,874 |
| Length | 127 |
| Max. P | 0.989671 |

| Location | 20,970,747 – 20,970,840 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 75.45 |
| Shannon entropy | 0.46525 |
| G+C content | 0.48274 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -15.31 |
| Energy contribution | -17.31 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
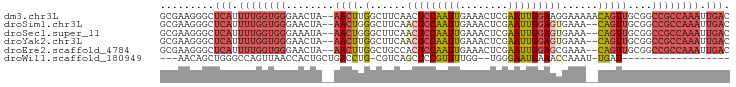
>dm3.chr3L 20970747 93 + 24543557 GCGAAGGGCUCAUUUUGGUGGGAACUA--AACUUGGCUUCAACUCCAAUUGAAACUCGAAUUGGAAGGAAAAACAGUUGCGGCCGCCAAAUUGAC ((.....))(((.(((((((....)..--.....((((.(((((((((((........))))))).(......).)))).)))))))))).))). ( -25.60, z-score = -1.49, R) >droSim1.chr3L 20325419 91 + 22553184 GCGAAGGGCUCAUUUUGGUGGGAACUA--AACUGGGCUUCAACUCCAAUUGAAACUCGAAUUGGAGUGAAA--CAGUUGCGGCCGCCAAAUUGAC ((.....))(((.((((((((......--(((((...((((.((((((((........)))))))))))).--)))))....)))))))).))). ( -32.00, z-score = -3.32, R) >droSec1.super_11 867722 91 + 2888827 GCGAAGGGCUCAUUUUGGUGGGAAAUA--AACUGGGCUUCAACUCCAAUUGAAACUCGAAUUGGAGUGAAA--CAGUUGCGGCCGCCAAAUUGAC ((.....))(((.((((((((......--(((((...((((.((((((((........)))))))))))).--)))))....)))))))).))). ( -32.00, z-score = -3.49, R) >droYak2.chr3L 4633434 91 - 24197627 GCGAAGGGCUCAUUUUGGUGGGAACUA--AACUUGGCUUCAACUCCAAUUGAAACUCGAAUUGGAGUGAAA--CAGUUGCGGCCGCCAAAUUGAC ((.....))(((.((((((((......--((((.(..((((.((((((((........)))))))))))).--)))))....)))))))).))). ( -29.10, z-score = -2.54, R) >droEre2.scaffold_4784 20659266 91 + 25762168 GCGAAGGGCUCAUUUUGGUGGGAACUA--AACUUGGCUGCCACUCCAAUUGAAACUCGAAUUGGAGCGAAA--CAGUUGCGGCCGCCAAAUUGAC ....((..(((((....)))))..)).--...(((((.(((..(((((((........)))))))((((..--...))))))).)))))...... ( -29.30, z-score = -2.00, R) >droWil1.scaffold_180949 936271 70 - 6375548 ---AACAGCUGGGCCAGUUAACCACUGCUGACCUG-CGUCAGCUCCGUUUUGG--UGGGAAUGAAACCAAAU-UGAU------------------ ---..(((.(((..((.....(((((((((((...-.))))))........))--)))...))...)))..)-))..------------------ ( -17.00, z-score = 0.21, R) >consensus GCGAAGGGCUCAUUUUGGUGGGAACUA__AACUUGGCUUCAACUCCAAUUGAAACUCGAAUUGGAGUGAAA__CAGUUGCGGCCGCCAAAUUGAC .........(((.((((((((........((((........(((((((((........))))))))).......))))....)))))))).))). (-15.31 = -17.31 + 2.00)
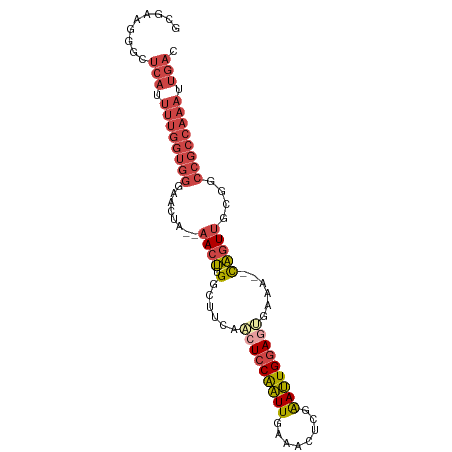
| Location | 20,970,747 – 20,970,840 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 75.45 |
| Shannon entropy | 0.46525 |
| G+C content | 0.48274 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -15.08 |
| Energy contribution | -16.58 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20970747 93 - 24543557 GUCAAUUUGGCGGCCGCAACUGUUUUUCCUUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCAAGUU--UAGUUCCCACCAAAAUGAGCCCUUCGC .(((.(((((.((..(((((((((((..((.((((((.(.....)))))))))..)))))..))))--..))..)).))))).)))......... ( -22.50, z-score = -1.53, R) >droSim1.chr3L 20325419 91 - 22553184 GUCAAUUUGGCGGCCGCAACUG--UUUCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCCAGUU--UAGUUCCCACCAAAAUGAGCCCUUCGC .(((.(((((.((..(((((((--(((((((((((((.(.....))))))))).))))))..))))--..))..)).))))).)))......... ( -27.90, z-score = -3.40, R) >droSec1.super_11 867722 91 - 2888827 GUCAAUUUGGCGGCCGCAACUG--UUUCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCCAGUU--UAUUUCCCACCAAAAUGAGCCCUUCGC .(((.(((((.((....(((((--(((((((((((((.(.....))))))))).))))))..))))--......)).))))).)))......... ( -26.40, z-score = -3.13, R) >droYak2.chr3L 4633434 91 + 24197627 GUCAAUUUGGCGGCCGCAACUG--UUUCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCAAGUU--UAGUUCCCACCAAAAUGAGCCCUUCGC .(((.(((((.((..(((((((--(((((((((((((.(.....))))))))).))))))..))))--..))..)).))))).)))......... ( -27.90, z-score = -3.34, R) >droEre2.scaffold_4784 20659266 91 - 25762168 GUCAAUUUGGCGGCCGCAACUG--UUUCGCUCCAAUUCGAGUUUCAAUUGGAGUGGCAGCCAAGUU--UAGUUCCCACCAAAAUGAGCCCUUCGC .(((.(((((.((..(((((((--(((((((((((((.(.....)))))))))))).)))..))))--..))..)).))))).)))......... ( -27.00, z-score = -2.00, R) >droWil1.scaffold_180949 936271 70 + 6375548 ------------------AUCA-AUUUGGUUUCAUUCCCA--CCAAAACGGAGCUGACG-CAGGUCAGCAGUGGUUAACUGGCCCAGCUGUU--- ------------------....-.((((((.........)--)))))((((.((((((.-...)))))).(.((((....)))))..)))).--- ( -18.70, z-score = -0.48, R) >consensus GUCAAUUUGGCGGCCGCAACUG__UUUCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCAAGUU__UAGUUCCCACCAAAAUGAGCCCUUCGC .(((.(((((.((....((((...(((((((((((((........))))))))).))))...))))........)).))))).)))......... (-15.08 = -16.58 + 1.51)

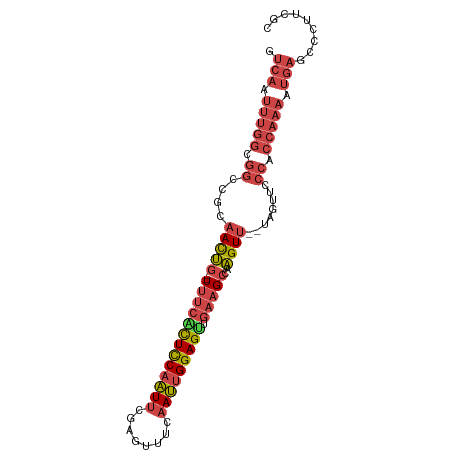
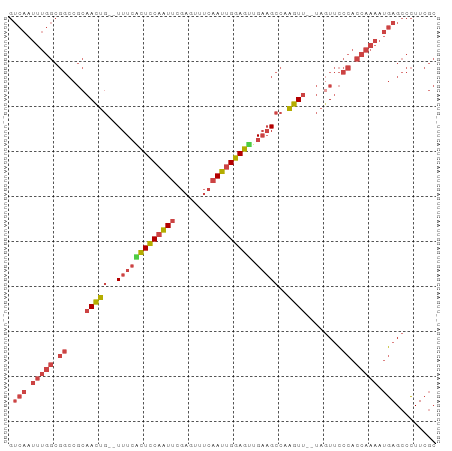
| Location | 20,970,762 – 20,970,874 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 96.30 |
| Shannon entropy | 0.06603 |
| G+C content | 0.50731 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -31.92 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20970762 112 - 24543557 GCCUGGGUGUGGCAUGCAACAGUCGACACGAGAAGUCAAUUUGGCGGCCGCAACUGUUUUUCCUUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCAAGUUUAGUUCCCACCA ...((((.(((((.(((...(((.(((.......))).)))..))))))))(((((((((..((.((((((.(.....)))))))))..)))))..)))).....))))... ( -30.40, z-score = -0.26, R) >droSim1.chr3L 20325434 110 - 22553184 GCCUGGGUGUGGCAUGCAACAGUCGACACGAGAAGUCAAUUUGGCGGCCGCAACUG--UUUCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCCAGUUUAGUUCCCACCA ..(((((((((((.(((...(((.(((.......))).)))..)))))))).....--.((((((((((((.(.....))))))))).)))))))))).............. ( -36.60, z-score = -1.94, R) >droSec1.super_11 867737 110 - 2888827 GCCUGGGUGUGGCAUGCAACAGUCGACACGAGAAGUCAAUUUGGCGGCCGCAACUG--UUUCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCCAGUUUAUUUCCCACCA ..(((((((((((.(((...(((.(((.......))).)))..)))))))).....--.((((((((((((.(.....))))))))).)))))))))).............. ( -36.60, z-score = -2.19, R) >droYak2.chr3L 4633449 110 + 24197627 GCCUGGGUGUGGCAUGCAACAGCCGACACGAGAAGUCAAUUUGGCGGCCGCAACUG--UUUCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCAAGUUUAGUUCCCACCA ....(((((((((........))).))))(.(((.(.((((((((....((....)--)((((((((((((.(.....))))))))).)))))))))))).).))).).)). ( -35.40, z-score = -1.52, R) >droEre2.scaffold_4784 20659281 110 - 25762168 GCCUGGGUGUGGCAUGCAACAGUCGACACGAGAAGUCAAUUUGGCGGCCGCAACUG--UUUCGCUCCAAUUCGAGUUUCAAUUGGAGUGGCAGCCAAGUUUAGUUCCCACCA ....(((((((((........))).))))(.(((.(.((((((((.(((((....)--)...(((((((((.(.....))))))))))))).)))))))).).))).).)). ( -37.50, z-score = -1.47, R) >consensus GCCUGGGUGUGGCAUGCAACAGUCGACACGAGAAGUCAAUUUGGCGGCCGCAACUG__UUUCACUCCAAUUCGAGUUUCAAUUGGAGUUGAAGCCAAGUUUAGUUCCCACCA ...((((.(((((.(((...(((.(((.......))).)))..))))))))((((...(((((((((((((.(.....)))))))))).))))...)))).....))))... (-31.92 = -32.24 + 0.32)

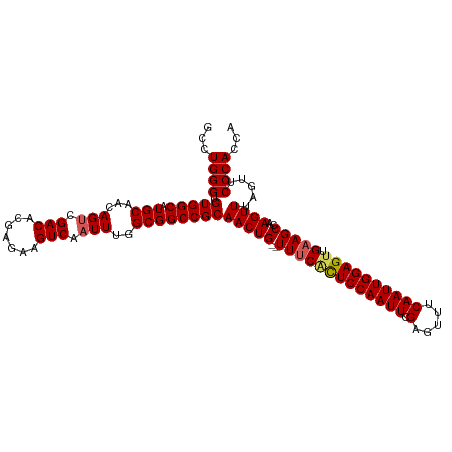
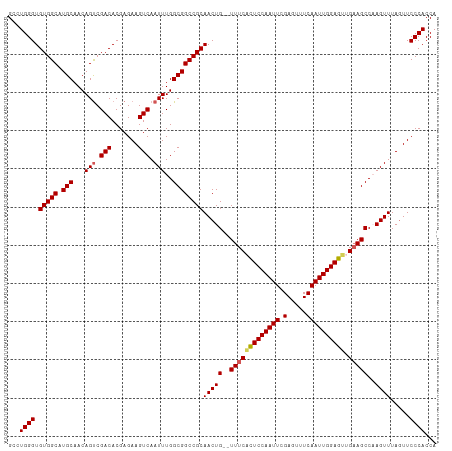
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:41:10 2011