| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,851,832 – 20,851,923 |
| Length | 91 |
| Max. P | 0.997094 |

| Location | 20,851,832 – 20,851,923 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 66.50 |
| Shannon entropy | 0.65025 |
| G+C content | 0.50903 |
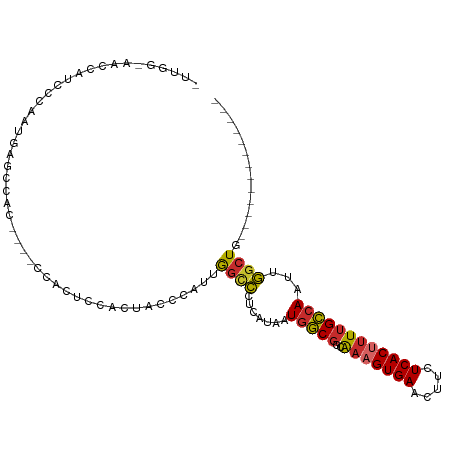
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -12.95 |
| Energy contribution | -12.90 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
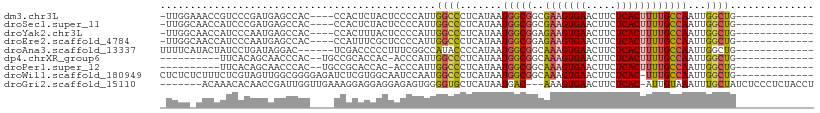
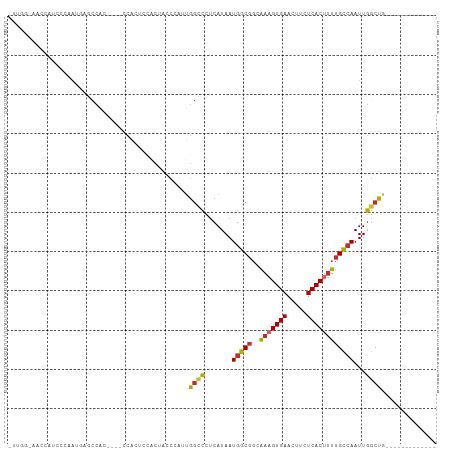
>dm3.chr3L 20851832 91 - 24543557 -UUGGAAACCGUCCCGAUGAGCCAC----CCACUCUACUCCCCAUUGGCCCUCAUAAUGGCGGCGAAGUGAACUUCUCACUUUUGCCAAUUGGCUG------------- -((((........))))..(((((.----............((((((.......)))))).((((((((((.....)))).))))))...))))).------------- ( -25.40, z-score = -2.19, R) >droSec1.super_11 747678 91 - 2888827 -UUGGCAACCAUCCCGAUGAGCCAC----CCACUCUACUCCCCAUUGGCCCUCAUAAUGGCGGCGAAGUGAACUUCUCACUUUUGCCAAUUGGCUG------------- -((((........))))..(((((.----............((((((.......)))))).((((((((((.....)))).))))))...))))).------------- ( -24.60, z-score = -1.97, R) >droYak2.chr3L 4509169 91 + 24197627 -UUGGCAACCAUCCCAAUGAGCCAC----CCACUUUACUCCCCAUUGGCCCUCAUAAUGGCGGAGAAGUGAACUUCUCACUUUUGCCAAUUGGCUG------------- -((((........))))..(((((.----........((((((((((.......)))))).))))((((((.....))))))........))))).------------- ( -25.50, z-score = -2.37, R) >droEre2.scaffold_4784 20538914 91 - 25762168 -UUGGCAACCAUCCCAAUGAGCCAC----CCAUUUCGCUCCCCAUUGGCCCUCAUAAUGGCGGAGAAGUGAACUUCUCACUUUUGCCAAUUGGCUG------------- -(((((((......((((((((...----.......))))...))))(((........))).((((((....))))))....))))))).......------------- ( -25.30, z-score = -1.67, R) >droAna3.scaffold_13337 14460882 90 + 23293914 UUUUCAUACUAUCCUGAUAGGAC------UCGACCCCCUUUCGGCCAUACCCCAUAAUGGCGGCAAAGUGAACUUCUCACUUUUGCCAAUUGGCUG------------- ...............((.(((..------.......))).))(((((....((.....)).((((((((((.....)))).))))))...))))).------------- ( -22.90, z-score = -2.64, R) >dp4.chrXR_group6 12025009 83 + 13314419 ----------UUCACAGCAACCCAC--UGCCGCACCAC-ACCCAUUGGCCCUCAUAAUGGCGGCAAAGUGAACUUCUCACUUUUGCCAAUUGGCUG------------- ----------....((((.......--...........-..((((((.......)))))).((((((((((.....)))).)))))).....))))------------- ( -21.80, z-score = -1.89, R) >droPer1.super_12 436033 83 + 2414086 ----------UUCACAGCAACCCAC--UGCCGCACCAC-ACCCAUUGGCCCUCAUAAUGGCGGCAAAGUGAACUUCUCACUUUUGCCAAUUGGCUG------------- ----------....((((.......--...........-..((((((.......)))))).((((((((((.....)))).)))))).....))))------------- ( -21.80, z-score = -1.89, R) >droWil1.scaffold_180949 781988 95 + 6375548 CUCUCUCUUUCUCGUAGUUGGCGGGGAGAUCUCGUGGCAAUCCAAUGGCCCUCAUAAUGGCGGCAAAGUGAACUUCUCAC-UUUGCCAAUUGGCUG------------- .(((((((.((........)).)))))))............(((((.(((........)))((((((((((.....))))-)))))).)))))...------------- ( -32.70, z-score = -2.67, R) >droGri2.scaffold_15110 2850632 98 - 24565398 -------ACAAACACAACCGAUUGGUUGAAAGGAGGAGGAGAGUGGGGUGCUCAUAAUGAC---AAAGUGAACUUCUCAC-AUUGUAAAUUUGCUAUCUCCCUCUACCU -------.......(((((....)))))..(((((..(((((((((((.(.((((......---...)))).).))))))-...(((....)))..)))))..)).))) ( -27.70, z-score = -1.58, R) >consensus _UUGG_AACCAUCCCAAUGAGCCAC____CCACUCCACUACCCAUUGGCCCUCAUAAUGGCGGCAAAGUGAACUUCUCACUUUUGCCAAUUGGCUG_____________ ..............................................((((.......(((((..(((((((.....))))))))))))...)))).............. (-12.95 = -12.90 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:59 2011