
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,847,151 – 20,847,321 |
| Length | 170 |
| Max. P | 0.864380 |

| Location | 20,847,151 – 20,847,321 |
|---|---|
| Length | 170 |
| Sequences | 15 |
| Columns | 174 |
| Reading direction | forward |
| Mean pairwise identity | 71.90 |
| Shannon entropy | 0.64476 |
| G+C content | 0.54899 |
| Mean single sequence MFE | -61.06 |
| Consensus MFE | -22.24 |
| Energy contribution | -21.84 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20847151 170 + 24543557 CUCACCUGUCGCACUGCAUAGUAGCAGGUGAGGUUGCGCGGAUAGAUGCCCGGAAAGUUGGGCGACUGCAGGCGACACGGCUUCUUGUCGCAGUCGCUGAAGAUCCUGGAGCAGUACGUUCCGGGUAUGAGGUCACCUGGAAAGCGGAAGGCAGAUGUCACAGUCAGAAA---- (((((((((..(........)..))))))))).((((...((((..(((((.(....(..((.((((.((((((((.((((.....))))..))))))......((((((((.....))))))))..)).)))).))..)....).)..))))..))))...).)))...---- ( -68.40, z-score = -1.75, R) >droPer1.super_12 429083 156 - 2414086 CGUACCUGGCGCACGGCAUAGUAGCAGGUCAGAUUCCGGGGAUAGAUGCCUGGGUAGUUGGGCGACUGCAGGCGACAGGACUUCUUGUCGCAGUCACUGAAGAUCCUCGAGCAGUAAGUGCCCGGAAUGAGAUCCCCUGUGUGUCAAAGAGAGAGA------------------ ......(((((((((((......)).((((..((((((((.((...((((..(....)..))))(((((..((((((((....)))))))).(((......)))......)))))..)).))))))))..))))...)))))))))..........------------------ ( -60.80, z-score = -1.76, R) >dp4.chrXR_group6 12018926 156 - 13314419 CGUACCUGGCGCACGGCAUAGUAGCAGGUGAGAUUCCGGGGAUAGAUGCCUGGGUAGUUGGGCGACUGCAGGCGACAGGACUUCUUGUCGCAGUCACUAAAGAUCCUCGAGCAGUAAGUGCCCGGAAUGAGAUCCCCUGUGUGUCAAAGAGAGAGA------------------ ......((((((((.((......))(((.((.((((((((.((...((((.((((..((((..((((((....((((((....)))))))))))).))))..))))..).)))....)).))))))))....)).)))))))))))..........------------------ ( -60.20, z-score = -1.92, R) >droAna3.scaffold_13337 14457053 174 - 23293914 CUCACCUGUCGCACUGCGUAGUAGCAGGUGAGGUUGCGGGGAUAGAUGCCGGGGAAGUUGGGCGACUGCAGGCGACAGGCCUUCUUGUCGCAGUCGCUGAAGAUCCUGGAGCAGUAGGUCCCCGGAAUAAGGUCACCUGCGAAUCAGAGUCAGAAUCAGACAAACACUUGCAAA (((((((((..(........)..))))))))).((((((((((...((((.((((..((.(((((((((....((((((....))))))))))))))).))..)))).).)))((((((..((.......))..))))))..)))...(((.......))).....))))))). ( -74.90, z-score = -3.21, R) >droYak2.chr3L 4504599 170 - 24197627 CUCACCUGUCGCACUGCGUAGUAGCAGGUGAGGUUGCGCGGAUAGAUGCCUGGGAAGUUGGGCGACUGCAGGCGACACGGCUUCUUGUCGCAGUCGCUGAAGAUCCUGGAGCAAUACGUUCCGGGUAUGAGGUCACCUGCAAAGCGGAAAGCAGAUGUCACAGCCAGAAA---- .....((((.(((((((...((.((((((((...(((.((((....((((..(((..((.(((((((((.(....)..(((.....)))))))))))).))..)))..).)))......)))).))).....))))))))...)).....)))).))).)))).......---- ( -70.90, z-score = -2.12, R) >droEre2.scaffold_4784 20534090 170 + 25762168 CUCACCUGUCGCACAGCGUAGUAGCAGGUGAGGUUGCGUGGAUAGAUGCCUGGGAAGUUGGGCGACUGCAGGCGACACGGCUUCUUGUCGCAGUCGCUGAAGAUCCUGGAGCAAUAUGUUCCGGGUAUGAGGUCACCUGCAAGGCGGGAAGCAGAUGUCCCAGCCAGAAA---- (((((((((..(........)..))))))))).(((((.((((..((((((((((..((.(((((((((.(....)..(((.....)))))))))))).))..))))(((((.....)))))))))))...))).).)))))(((((((........)))).))).....---- ( -76.40, z-score = -3.23, R) >droSec1.super_11 742869 170 + 2888827 CUCACCUGUCGCACUGCGUAGUAGCAGGUGAGGUUGCGCGGAUAGAUGCCCGGAAAGUUGGGCGACUGCAGGCGACACGGCUUCUUGUCGCAGUCGCUGAAGAUCCUGGAGCAGUACGUUCCGGGAAUGAGGUCACCUGGAAAGCGGAAGGCAGAUGUCACAGCCAGAAA---- (((((((((..(........)..))))))))).(((.((.((((..(((((.(....(..((.((((.((((((((.((((.....))))..)))))).....(((((((((.....))))))))).)).)))).))..)....).)..))))..))))...)))))...---- ( -73.30, z-score = -2.61, R) >droSim1.chr3L_random 890925 170 + 1049610 CUCACCUGUCGCACUGCGUAGUAGCAGGUGAGGUUGCGCGGAUAGAUGCCCGGAAAGUUGGGCGACUGCAGGCGACACGGCUUCUUGUCGCAGUCGCUGAAGAUCCUGGAGCAGUACGUUCCGGGUAUGAGGUCACCUGGAAAGCGAAAGGCAGAUGUCACAGCCAGAAA---- (((((((((..(........)..))))))))).(((.((.((((..((((((.....(..((.((((.((((((((.((((.....))))..))))))......((((((((.....))))))))..)).)))).))..)....))...))))..))))...)))))...---- ( -69.90, z-score = -1.93, R) >droWil1.scaffold_180949 777377 171 - 6375548 CCUACCUGUCGCACCGCAUAGUAGCAAGUAAGAUUACGUGGAUAGAUGCCAGGGAAAUUGGGUGAUUGCAGGCGACACGACUUCUUGUCGCAAUCACUGAAUAUGCGUGAGCAAUAUGUGCCAGGAAUGAGAUCACCUAAACCAAAGGAAUACGAAUAUAGAAUACAUAAA--- (((..(((((.(((.((......))..(((....)))))))))))......((....(((((((((..((.((((((.(....).))))))..((.(((.((((((....)))...)))..))))).))..))))))))).))..))).......................--- ( -46.60, z-score = -1.76, R) >droVir3.scaffold_13049 1777169 149 + 25233164 CUUACCUGUCGCACCGCAUAGUAGCAGGUGAGAUUGCGCGGAUAGAUGCCCGGAUAGUUGGGCGAUUGCAGGCGGCAGGGCUUCUUGUCGCAAUCGCUGAAGAUGCGCGAGCAAUAGGUGCCCGGUAUUAGGUCGCCUGUGAAGUGGGG------------------------- (((((((((..(........)..)))))))))....(((.......(((((((....))))))).(..((((((((.((((.(((((((((.(((......)))..)))).))..))).))))........))))))))..).)))...------------------------- ( -61.30, z-score = -1.49, R) >droMoj3.scaffold_6654 508797 142 - 2564135 CCUACCUGUCGCACUGCAUAAUAGCAGGUGAGAUUGCGCGGAUAGAUGCCCGGAUAGUUGGGUGACUGCAAGCGGCAGGGCUUCUUGUCGCAGUCGCUAAAGAUGCGCGAGCAGUAGGUGCCUGGUAUAAGAUCACCUGAAA-------------------------------- ....((((((((.((((......))))..........((((.....(((((((....))))))).))))..))))))))(((.((((.((((.((......)))))))))).)))(((((.((......))..)))))....-------------------------------- ( -50.40, z-score = -0.40, R) >droGri2.scaffold_15110 2847436 149 + 24565398 CUCACCUGUCGCACCGCAUAGUAGCAGGUGAGAUUGCGCGGAUAGAUGCCCGGAUAGUUGGGCGAUUGCAGGCGACACGGCUUCUUGUCGCAAUCGCUGAAGAUUCUCGAACAAUAUGUGCCCGGGAUGAGGUCGCCUGUGCAAAAGAU------------------------- (((((((((..(........)..))))))))).((((((((...(((.(((((....((.(((((((((.(....)..(((.....)))))))))))).)).........((.....))..))))).....)))..)))))))).....------------------------- ( -58.30, z-score = -1.96, R) >anoGam1.chr2L 17212478 155 - 48795086 CUUACCUGUCUAACGGCGAAAUAGCACGUCAGAUUCCUGGGAUAGAUGCCGGGAAAGUUGGGCGACUGCAGACGGCACACCUUUUUGUCGCAGUUGGUGAAUAUACGCGAGCAGAACGUACCCUGCAUGAGAUCGCCU--GCGGAAAUAGUACGAAA----------------- .(((((.(((((((((((........))))...(((((((........))))))).)))))))((((((.(((((.........))))))))))))))))................(((((.(((((.(......).)--)))).....)))))...----------------- ( -47.70, z-score = -0.58, R) >triCas2.ChLG9 11958814 165 + 15222296 -----CUGACGCACUGCGUAAUAACAGGUUAAAUUGCGCGGGUAAACCCCCGGAAAGUUGGGGGACUGGAGACGGCAGUGCUUCCUAUCACAGUCGCUGAAAAUUCGGGAACAAUAUGUACCCGUUAUUAAAUCUCCCAAGUAGUAUUCGGGUUCGGGCUCAACUGAGGG---- -----((((.((.(((((((((..........))))))))))).....((((((.(.(((((((((((....)))(((((((.........)).)))))......((((.((.....)).))))........)))))))).)....)))))).)))).(((....)))..---- ( -51.60, z-score = -0.66, R) >apiMel3.Group11 10768394 128 - 12576330 ------------ACUGCGUAGUAGCAGGUGAGAUUGCGCGGAUAUAAUCCUGGAAAGUUGGGCGACUGGAGCCUACACUUUUUCCGGUCGCAGUCGCUGAAUAUGCGGGAACAGUAGGUGCCCGAUAUCAGAUCCCCCAG---------------------------------- ------------.(((((((((..(....)..)))))))))........((((........((((((((((..........)))))))))).....((((.(((.((((.((.....)).))))))))))).....))))---------------------------------- ( -45.20, z-score = -1.35, R) >consensus CUCACCUGUCGCACUGCGUAGUAGCAGGUGAGAUUGCGCGGAUAGAUGCCCGGAAAGUUGGGCGACUGCAGGCGACACGGCUUCUUGUCGCAGUCGCUGAAGAUCCUGGAGCAGUACGUGCCCGGUAUGAGGUCACCUGGAAAGCAGAAAGAAGAU_____A____________ .........(((...((......))..(........))))(((.....((.((....((.(((((((((.(((((.........)))))))))))))).)).........((.....)).)).))......)))........................................ (-22.24 = -21.84 + -0.40)

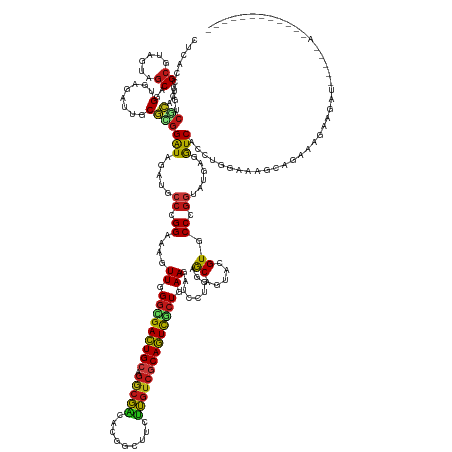
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:57 2011