
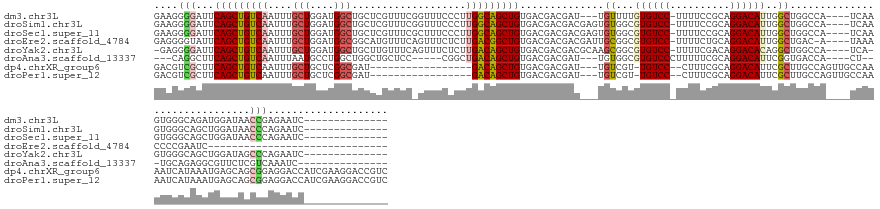
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,818,402 – 20,818,539 |
| Length | 137 |
| Max. P | 0.859683 |

| Location | 20,818,402 – 20,818,539 |
|---|---|
| Length | 137 |
| Sequences | 8 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 65.05 |
| Shannon entropy | 0.65971 |
| G+C content | 0.54158 |
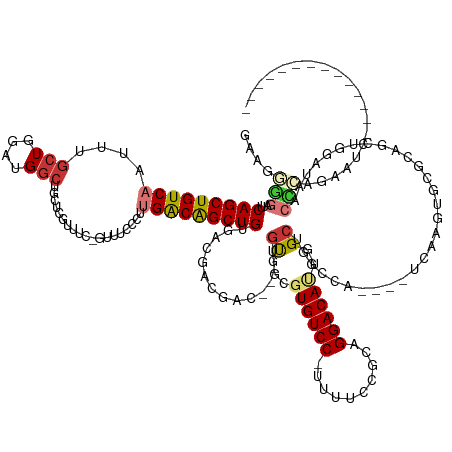
| Mean single sequence MFE | -51.38 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.67 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20818402 137 + 24543557 GAAGGGGAUUCAGCUGUCAAUUUGCUGGAUGGCUGCUCGUUUCGGUUUCCCUUGGCAGCUGUGACGACGAU---UGUUUUGUGUCC-UUUUCCGCAGGACAUUGGCUGGCCA----UCAAGUGGGCAGAUGGAUAACCGAGAAUC-------------- ...((..(((((.(((((.....(((.((((((((((((((.(((((.((...)).))))).)))))((((---.((((((((...-.....)))))))))))))).)))))----)).))).))))).)))))..)).......-------------- ( -51.40, z-score = -1.95, R) >droSim1.chr3L 20173976 140 + 22553184 GAAGGGGAUUCAGCUGUCAAUUUGCUGGAUGGCUGCUCGUUUCGGUUUCCCUUGGCAGCUGUGACGACGACGAGUGUGGCGUGUCC-UUUUCCGCAGGACAUUGGCUGGCCA----UCAAGUGGGCAGCUGGAUAACCCAGAAUC-------------- ...(((.(((((((((((.....(((.((((((((((((((.(((((.((...)).))))).......)))))))..(((((((((-(.......)))))))..))))))))----)).))).)))))))))))..)))......-------------- ( -57.20, z-score = -2.05, R) >droSec1.super_11 714239 140 + 2888827 GAAGGGGAUUCAGCUGUCAAUUUGCUGGAUGGCUGCUCGUUUCGCUUUCCCUUGGCAGCUGUGACGACGACGAGUGUGGCGUGUCC-UUUUCCGCAGGACAUUGGCUGGCCA----UCAAGUGGGCAGCUGGAUAACCCAGAAUC-------------- ...(((.(((((((((((.....(((.((((((((((((((((((....(....).....))))....)))))))..(((((((((-(.......)))))))..))))))))----)).))).)))))))))))..)))......-------------- ( -56.20, z-score = -1.77, R) >droEre2.scaffold_4784 20505693 123 + 25762168 GAGGGGUAUUCAGCUGUCAAUUUGCUGGAUGGCGGCAUGUUUCAGUUUCUCUUGACGGCUGUGACGACGACGAUUGCGGCGUGUCC-UUUUCUGCAGGACAUUGGCUGAC-A----UAAACCCCGAAUC------------------------------ ..(((((...((((((((((...((((((...........)))))).....)))))))))).............((((((((((((-(.......)))))))..)))).)-)----...))))).....------------------------------ ( -42.60, z-score = -1.62, R) >droYak2.chr3L 4476217 138 - 24197627 -GAGGGGAUUCAGCUGUCAAUUUGCUGGAUGGCUGCUUGUUUCAGUUUCUCUUGACAGCUGUGACGACGACGCAAGCGGCGUGUCC-UUUUCGACAGGACACAGGCUGGCCA----UCA-GUGGGCAGCUGGAUAGCCCAGAAUC-------------- -..(((.(((((((((((.....((((.(((((((((((((.(((((((....)).))))).)))..((.(....)))..((((((-(.......))))))))))).)))))----)))-)).)))))))))))..)))......-------------- ( -60.70, z-score = -3.42, R) >droAna3.scaffold_13337 14428999 126 - 23293914 ---CAGGCUUCAGCUGUCAAUUUAAUGCCUGGCUGGCUGCUCC-----CGGCUGACAGCUGUGACGACGAU---UGUGGCGUGUCCCUUUUUCGCAGGACAUUCGGUGACCA----CU---UGCAGAGGCGUUCUCGUCAAAUC--------------- ---.......(((((((((.......(((.....))).((...-----..)))))))))))((((((.((.---(((.((((((((..........))))))..(....)..----..---.))....))).))))))))....--------------- ( -39.30, z-score = 0.53, R) >dp4.chrXR_group6 11983466 136 - 13314419 GACGUCGCUUCAGCUGUCAAUUUGCUGCUCGGCGAU-----------------GACAGCUGUGACGACGAU---UGUCGU-UGUCC--CUUUCGCAGGACAUUCGCUUGCCAGUUGCCAAAAUCAUAAAUGAGCAGCGGAGGACCAUCGAAGGACCGUC ((((((.((((....(((..(((((((((((...((-----------------((((((((.(((((((..---...)))-)))).--.....(((((.(....))))))))))))......))))...))))))))))).)))....)))))).)))) ( -51.80, z-score = -2.41, R) >droPer1.super_12 394005 136 - 2414086 GACGUCGCUUCAGCUGUCAAUUUGCUGCUCGGCGAU-----------------GACAGCUGUGACGACGAU---UGUCGU-UGUCC--CUUUCGCAGGACAUUCGCUUGCCAGUUGCCAAAAUCAUAAAUGAGCAGCGGAGGACCAUCGAAGGACCGUC ((((((.((((....(((..(((((((((((...((-----------------((((((((.(((((((..---...)))-)))).--.....(((((.(....))))))))))))......))))...))))))))))).)))....)))))).)))) ( -51.80, z-score = -2.41, R) >consensus GAAGGGGAUUCAGCUGUCAAUUUGCUGGAUGGCUGCUCGUUUC_GUUUCCCUUGACAGCUGUGACGACGAC___UGUGGCGUGUCC_UUUUCCGCAGGACAUUGGCUGGCCA____UCAAGUGCGCAGCUGGAUAACCCAGAAUC______________ ....(((...(((((((((...........(((.....)))...........)))))))))..............((...((((((..........))))))..))..............................))).................... (-16.80 = -17.67 + 0.88)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:55 2011