
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,772,277 – 20,772,388 |
| Length | 111 |
| Max. P | 0.549664 |

| Location | 20,772,277 – 20,772,388 |
|---|---|
| Length | 111 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.37 |
| Shannon entropy | 0.64402 |
| G+C content | 0.52040 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -15.14 |
| Energy contribution | -14.50 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.19 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20772277 111 + 24543557 CGGUCCAAAGGUCUCACCGCUCACGAAAUUCAGCUGGCCUGUGAACGGGCCGGCGUUUUCAC---CCAAGACCCCAACAAG------CCCAAUCCGAAUCCGAACACCGUCAUUAGCAUU ((((.....(((((....(....)((((....(((((((((....)))))))))..))))..---...)))))........------.......((....))...))))........... ( -29.80, z-score = -1.13, R) >dp4.chrXR_group8 8194270 99 + 9212921 CGGUCCAAAGGUCUGACGGCCCAUGAAAUACAGUUGGCUUGCGAGAGGGCCGGCGUUUUUAC---CCAAGAUCCGAAUAAU------CCCAACACGGUUAUCAACAUU------------ (((((....((((....))))..(((((....((((((((......))))))))..))))).---....)).))).(((((------(.......)))))).......------------ ( -20.80, z-score = 1.43, R) >droPer1.super_40 385238 99 + 810552 CGGUCCAAAGGUCUGACGGCCCAUGAAAUACAGUUGGCUUGCGAGAGGGCCGGCGUUUUUAC---CCAAGAUCCGAAUAAU------CCCAACACGGUUAUCAACAUU------------ (((((....((((....))))..(((((....((((((((......))))))))..))))).---....)).))).(((((------(.......)))))).......------------ ( -20.80, z-score = 1.43, R) >droSim1.chr3L 20128323 111 + 22553184 CGGUCCAAAGGUCUCACCGCUCACGAAAUCCAGCUGGCCUGUGAACGGGCCGGCGUUUUCAC---UCAAGACCCAAACAAU------CCCAAUCCCAGUCCGAACACCGUCAUUAGCAUU .((((....((.....))......((((....(((((((((....)))))))))..))))..---....))))........------................................. ( -29.40, z-score = -1.49, R) >droSec1.super_11 671429 111 + 2888827 CGGUCCAAAGGUCUCACCGCUCACGAAAUCCAGCUGGCCUGUGAACGGGCCGGCGUUUUCAC---CCAAGACCCCAACAAU------CCCAAUUCCAGUCCGAACACCGUCAUUAGCAUU .((((....((.....))......((((....(((((((((....)))))))))..))))..---....))))........------................................. ( -29.40, z-score = -1.42, R) >droYak2.chr3L 4419980 111 - 24197627 CGGUCCAAAGGUCUCACUUCUCACGAAAUCCAGUUAGCCUGCGAACGGGCCGGCGUUUUCAC---CCAAGACCCCAAUAAU------CCCAAUCCCAGCCCGAACACCGUGAUCAGCAUU .(..(....)..)...((..(((((...(((((.....))).)).(((((.((.(((.....---................------...))).)).))))).....)))))..)).... ( -20.26, z-score = 0.71, R) >droEre2.scaffold_4784 20463168 111 + 25762168 CGGUCCAAAGGUCUAACCGCUCACGAAAUCCAGUUGGCCUGCGAACGGGCCGGCGUUUUCAC---CCAAGACCCCAAUAAU------CUCAGUUCCAGUCCGAACACCGUGAUUAGCAUU .((((....((.....))......((((....(((((((((....)))))))))..))))..---....))))....((((------(.(.((((......))))...).)))))..... ( -29.50, z-score = -0.98, R) >droAna3.scaffold_13337 11126533 117 + 23293914 CGGUCCAAAGGUCUGACUGCCCACGAGAUCCAGCUGGCCUGCGAGCGGGCCGGCGUCUUCAC---CCAGGAUCCCAAUAAUUUUAAUCCGAAUUCCAAUCCCAAUACGGUCAUCAGCAUC .........(((((((..(((...(.(((((.(((((((((....)))))))))(....)..---...))))).)...(((((......))))).............)))..)))).))) ( -32.30, z-score = -0.31, R) >droMoj3.scaffold_6680 12675139 99 + 24764193 AGGUCCAAAGGUCUAACUGCGCAUGAGAUACAAUUGGCUUGUGAGCGAGCCGGUGUCUUUAC---CCAAGAUCCAAACAAU------CCCAAUACUGUCAUAAAUAUU------------ .((......(((((...((.(...(((((((....((((((....)))))).)))))))...---))))))))........------))...................------------ ( -20.24, z-score = -0.08, R) >droGri2.scaffold_15110 5516734 99 - 24565398 CGGUCCAAAGGUCUAACAGCUCAUGAGAUACAGCUGGCAUGCGAGCGGGCCGGUGUCUUUAG---CCAAGAUCCAAACAAU------CCAAAUACCGUUAUCAACAUU------------ .((......(((((....(((...(((((((.(((.((......)).)))..))))))).))---)..)))))........------))...................------------ ( -22.74, z-score = -0.26, R) >droVir3.scaffold_13049 6660993 99 - 25233164 CGGUCCAAAGGUCUAACUGCGCAUGAGAUACAAUUGGCUUGCGAGCGGGCCGGUGUCUUCAG---CCAAGAUCCAAACAAU------CCCAAUACCGUUAUCAAUAUU------------ ((((.....(((((...((.((.((((((((....((((((....)))))).))))).))))---))))))))........------......))))...........------------ ( -23.21, z-score = -0.06, R) >droWil1.scaffold_181024 1634621 99 + 1751709 CAGUCCAAAGGUCUCACGGCCCAUGAAAUACAAUUGGCAUGCGAGCGAGCCGGUGUUUUCAC---ACAAGAUCCAAGUAAU------CCCAAUACUGUAAUUCAAAUC------------ .........((((....))))..((((.(((((((((..(((..(.((....((((....))---))....)))..)))..------.)))))..)))).))))....------------ ( -19.90, z-score = -0.53, R) >anoGam1.chr2L 20799414 96 - 48795086 CGCUCCAAAGGACUGUCGGAGGAUGAGAUACAGAUUGCGUGCGAGCGAGCGGGCGUCUUUACG--ACCAGCCCCACACAAC----------AGACGGUCAUCAGCAUG------------ .(((......((((((((..(..((.......(.((((......)))).)(((((((.....)--))..))))))..)..)----------.)))))))...)))...------------ ( -27.00, z-score = 0.16, R) >triCas2.ChLG9 9801558 94 + 15222296 AAACGGAAGGGUUUAACCGAUGAGGAGAUCAGACAGGCGUGCGAGAAAUCGGGGGCUUACAAAUACCACGAACAAAGAAAU-------CGGACGCCGCCUC------------------- ...(....)(((....((.....))..........(((((.(((....(((.((...........)).))).........)-------)).))))))))..------------------- ( -22.12, z-score = -0.11, R) >consensus CGGUCCAAAGGUCUAACCGCUCAUGAAAUACAGUUGGCCUGCGAGCGGGCCGGCGUUUUCAC___CCAAGAUCCAAACAAU______CCCAAUACCGUUACCAACAUC____________ .........(((((..........(((((...((((((((......))))))))))))).........)))))............................................... (-15.14 = -14.50 + -0.64)

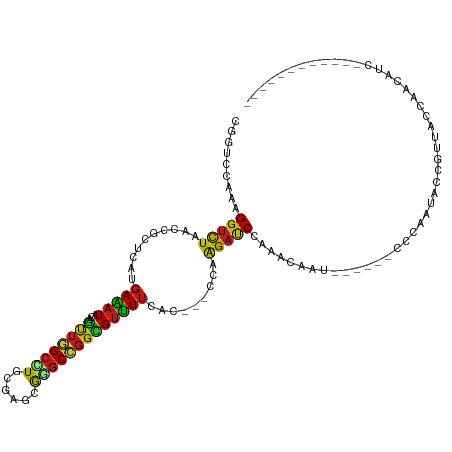

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:51 2011