
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,764,553 – 20,764,635 |
| Length | 82 |
| Max. P | 0.585639 |

| Location | 20,764,553 – 20,764,635 |
|---|---|
| Length | 82 |
| Sequences | 12 |
| Columns | 89 |
| Reading direction | forward |
| Mean pairwise identity | 62.14 |
| Shannon entropy | 0.79714 |
| G+C content | 0.49294 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -8.43 |
| Energy contribution | -8.43 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20764553 82 + 24543557 -UAGCAGCUGAUGCAUCAGCUGCUAGCACUGAAU-----UUCACGACGAGUUGGCAACACAAAGCCGAUGCGACGCCAUCUUGCUUUA- -((((((((((....))))))))))(((..((..-----....((.((.((((((........)))))).)).))...)).)))....- ( -28.20, z-score = -2.56, R) >droSim1.chr3L 20120562 82 + 22553184 -CAGCAGCUGAUGCAUCAGCUGCUAGCACUGAAU-----UUCUCGCAGAGUUGGCAGCACGAAACCGAUGCGACGCCAUCUUGCUUUG- -.(((((((((....)))))))))..........-----.....(((.((.((((.(((((....)).)))...)))).)))))....- ( -29.50, z-score = -1.64, R) >droSec1.super_11 663714 82 + 2888827 -CAGCAGCUGACGCAUCAGCUGGUAGCACUGAAU-----UUCUCGCCGAGUUGGCAGCACGAACCCGAUGCGACGCCAUCUUGCUUUA- -...(((((((....)))))))............-----.....((.(((.((((.(((((....)).)))...)))).)))))....- ( -24.50, z-score = -0.27, R) >droYak2.chr3L 4412233 81 - 24197627 -CAGCAGCUGACGCAUCAGCUGCUAGCACCGAAU-----UUAUCGCCGAGUUGGCAGU-CCAAGCUGAUGCGACGCCAUUUUGCUUAA- -.(((((((((....)))))))))((((.(((..-----...)))......((((.((-(...((....)))))))))...))))...- ( -26.80, z-score = -1.36, R) >droEre2.scaffold_4784 20455354 82 + 25762168 -CAGCAGCUGACGCAUCAGCUGCUAGCACUGAAU-----UCUUCGCCGAGUUGGCAGCACCGCGCUGAUGCGACGCCAUUUUGCUUUA- -.(((((((((....)))))))))((((......-----.....(((.....))).((..((((....))))..)).....))))...- ( -26.90, z-score = -0.32, R) >droAna3.scaffold_13337 11118502 78 + 23293914 -CAGCAGCUGAUACAACAGCUGUUAG----GAAA-----CUCGAACCAGGUUGGCAGACCCAAGCCGAUGCGACGCCAUUUUCUUUUC- -.((((((((......)))))))).(----(...-----.(((.....((((((.....)).))))....)))..))...........- ( -20.90, z-score = -1.41, R) >dp4.chrXR_group8 8187002 78 + 9212921 -UAUCAGCUGACGCAACAGCUGUUCGAAACAAA--------CGAGCCGAGUUGGCAGUGCCAAGC--GUGCGACGCCAUCUUUUCUUUA -....(((((......)))))(((((.......--------))))).(((.((((..((((....--).)))..)))).)))....... ( -20.00, z-score = 0.16, R) >droPer1.super_40 377962 78 + 810552 -UAGCAGCUGACGCAACAGCUGUUCGAAACAAA--------CGAGCCGAGUUGGCAGUGCCAAGC--GUGCGACGCCAUCUUUUCUUUA -.((((((((......)))))))).........--------.(((..(((.((((..((((....--).)))..)))).)))..))).. ( -22.10, z-score = -0.05, R) >droWil1.scaffold_180949 3851577 78 - 6375548 -AAUGAGUUGCUACAACAGCUGUUAAAUAAAAA--------UAAAACAAGUUGGCAUCACCAAACUGGU-UGUCGCCAUUUUUUUUCC- -....(((((......)))))............--------..........((((..((((.....)).-))..))))..........- ( -12.70, z-score = -0.25, R) >droMoj3.scaffold_6680 12062100 80 - 24764193 UAGUCAGCUGAGUGAACAGCUGG-AUCUUAACU-------UUCAAAUGAUGUGGCAACGUCUAGCAGACGCCAUUUUUUCCUGUCUUA- ...(((((((......)))))))-.........-------.......((.(((((...(((.....))))))))....))........- ( -17.10, z-score = 0.07, R) >droGri2.scaffold_15110 8930975 88 - 24565398 CAGUCAGCUGUGGCAACAGCUGUCAGCUUGAUUAGUUAUUUUCCAGCAAAGUGGCAACGUUGGACAGACGCCAUUUUUUCCUGUGUUG- ..(.(((((((....))))))).).(((.((..(....)..)).)))((((((((..(........)..))))))))...........- ( -24.30, z-score = -0.69, R) >droVir3.scaffold_13049 18545605 80 + 25233164 ----CAGCUGUGGCAACAGCUGGCAGUUAAAUUU----CAAUAUGACAAUAUGGCAACGUCGAGCAGACGCCAUUUUUUCCUGUCUUA- ----(((((((....)))))))((((..(((..(----(.....))....(((((...(((.....)))))))).)))..))))....- ( -21.80, z-score = -1.54, R) >consensus _CAGCAGCUGACGCAACAGCUGUUAGCACUAAAU_____UUCUCGCCGAGUUGGCAGCACCAAGCCGAUGCGACGCCAUCUUGUUUUA_ ....((((((......))))))................................................................... ( -8.43 = -8.43 + 0.01)

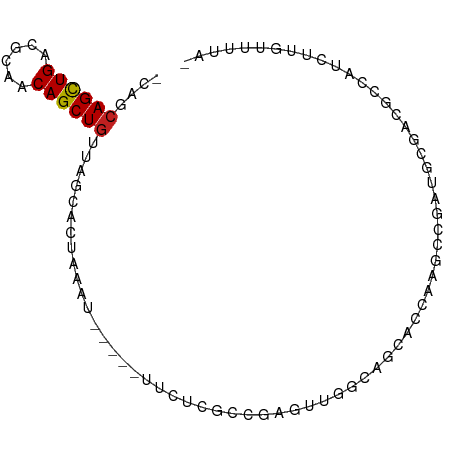
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:49 2011