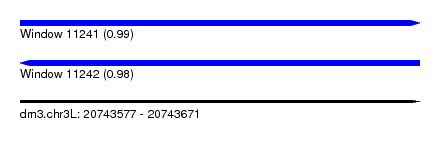
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,743,577 – 20,743,671 |
| Length | 94 |
| Max. P | 0.989767 |

| Location | 20,743,577 – 20,743,671 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 127 |
| Reading direction | forward |
| Mean pairwise identity | 62.97 |
| Shannon entropy | 0.62756 |
| G+C content | 0.56140 |
| Mean single sequence MFE | -38.93 |
| Consensus MFE | -15.57 |
| Energy contribution | -17.11 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
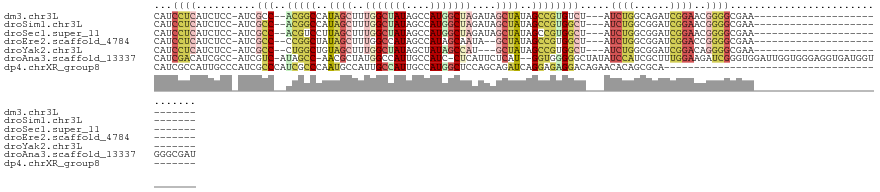
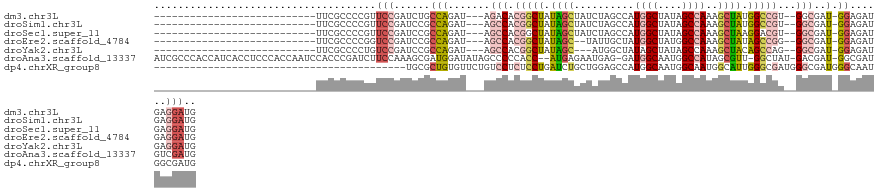
>dm3.chr3L 20743577 94 + 24543557 CAUCCUCAUCUCC-AUCGCC--ACGGCCAUAGCUUUGGCUAUAGCCAUGGCUAGAUAGCUAUAGCCGUGUCU---AUCUGGCAGAUCGGAACGGGGCGAA--------------------------- ..(((((...(((-((((((--(.(((((......)))))((((.((((((((........)))))))).))---)).)))).))).)))...))).)).--------------------------- ( -36.40, z-score = -2.30, R) >droSim1.chr3L 20100096 94 + 22553184 CAUCCUCAUCUCC-AUCGCC--ACGGCCAUAGCUUUGGCUAUAGCCAUGGCUAGAUAGCUAUAGCCGUGGCU---AUCUGGCGGAUCGGAACGGGGCGAA--------------------------- ..(((((...(((-.(((((--(.(((((......)))))(((((((((((((........)))))))))))---)).))))))...)))...))).)).--------------------------- ( -43.60, z-score = -3.68, R) >droSec1.super_11 632835 94 + 2888827 CAUCCUCAUCUCC-AUCGCC--ACGUCCUUAGCUUUGGCUAUAGCCAUGGCUAGAUAGCUAUAGCCGUGGCU---AUCUGGCGGAUCGGAACGGGGCGAA--------------------------- .............-.(((((--.(((((.......((((....))))..(((((((((((((....))))))---))))))).....)).))).))))).--------------------------- ( -40.10, z-score = -3.06, R) >droEre2.scaffold_4784 20436209 92 + 25762168 CAUCCUCAUCUCC-AUCGCC--CCGGCUAUAGCUUUGGCCAUAGCCAUAGCAAUA--GCUAUAGCCGUGGCU---AUCUGGCGGAUCGGACCGGGGCGAA--------------------------- .............-.(((((--((((((((.(((.((((((..((.(((((....--))))).))..)))))---)...)))..)).)).))))))))).--------------------------- ( -44.50, z-score = -4.01, R) >droYak2.chr3L 4394041 91 - 24197627 CAUCCUCAUCUCC-AUCGCC--CUGGCUGUAGCUUUGGCUAUAGCUAUAGCCAU---GCUAUAGCCGUGGCU---AUCUGGCGGAUCGGACAGGGGCGAA--------------------------- .............-.(((((--(.(((((((((..(((((((....))))))).---)))))))))....((---.(((((....))))).)))))))).--------------------------- ( -41.70, z-score = -3.13, R) >droAna3.scaffold_13337 11099341 121 + 23293914 CAUCGACAUCGCC-AUCGUC-AUAGCC-AACGCUAUGGCCAUUGCCAUC-CUCAUUCUCAU--GGUGGGGGCUAUAUCCAUCGCUUUGGAAGAUCGGGUGGAUUGGUGGGAGGUGAUGGUGGGCGAU .((((...(((((-((((.(-(((((.-...))))))((((...(((((-(...(((.((.--((((((((.....))).))))).)))))....))))))..))))......))))))))).)))) ( -47.50, z-score = -1.37, R) >dp4.chrXR_group8 8173019 85 + 9212921 CAUCGCCAUUGCCCAUCGCCCAUCGCCCAAUGCCAUUGCCAUUGCCAUGGCUCCAGCAGAUCAGGAGAGGACAGAACACAGCGCA------------------------------------------ ...(((..(((.((.((.((.(((((.....(((((.((....)).)))))....)).)))..)).)))).)))......)))..------------------------------------------ ( -18.70, z-score = 0.93, R) >consensus CAUCCUCAUCUCC_AUCGCC__ACGGCCAUAGCUUUGGCUAUAGCCAUGGCUAGAUAGCUAUAGCCGUGGCU___AUCUGGCGGAUCGGAACGGGGCGAA___________________________ ...((((..........(((..(((((..((((..(((((((....)))))))....))))..)))))))).....((((......))))..))))............................... (-15.57 = -17.11 + 1.54)
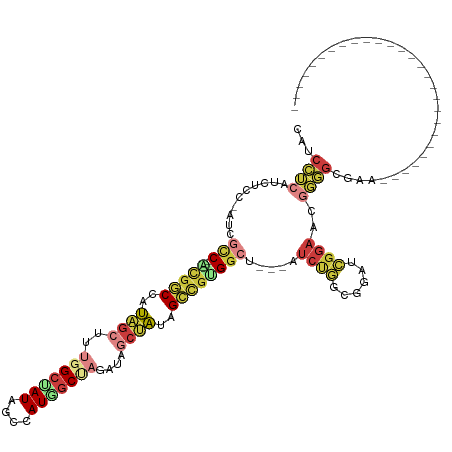
| Location | 20,743,577 – 20,743,671 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 127 |
| Reading direction | reverse |
| Mean pairwise identity | 62.97 |
| Shannon entropy | 0.62756 |
| G+C content | 0.56140 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -12.56 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20743577 94 - 24543557 ---------------------------UUCGCCCCGUUCCGAUCUGCCAGAU---AGACACGGCUAUAGCUAUCUAGCCAUGGCUAUAGCCAAAGCUAUGGCCGU--GGCGAU-GGAGAUGAGGAUG ---------------------------.......(((((..((((.(((..(---(((...(((....))).))))((((((((((((((....)))))))))))--)))..)-))))))..))))) ( -40.70, z-score = -3.75, R) >droSim1.chr3L 20100096 94 - 22553184 ---------------------------UUCGCCCCGUUCCGAUCCGCCAGAU---AGCCACGGCUAUAGCUAUCUAGCCAUGGCUAUAGCCAAAGCUAUGGCCGU--GGCGAU-GGAGAUGAGGAUG ---------------------------....((.(((..(.(((.((...((---(((....))))).))......((((((((((((((....)))))))))))--))))))-.)..))).))... ( -43.50, z-score = -4.31, R) >droSec1.super_11 632835 94 - 2888827 ---------------------------UUCGCCCCGUUCCGAUCCGCCAGAU---AGCCACGGCUAUAGCUAUCUAGCCAUGGCUAUAGCCAAAGCUAAGGACGU--GGCGAU-GGAGAUGAGGAUG ---------------------------....((.(((..(.(((.(((((.(---(((...((((((((((((......))))))))))))...))))....).)--))))))-.)..))).))... ( -36.00, z-score = -2.49, R) >droEre2.scaffold_4784 20436209 92 - 25762168 ---------------------------UUCGCCCCGGUCCGAUCCGCCAGAU---AGCCACGGCUAUAGC--UAUUGCUAUGGCUAUGGCCAAAGCUAUAGCCGG--GGCGAU-GGAGAUGAGGAUG ---------------------------.((((((((((..(......)..((---(((...(((((((((--(((....))))))))))))...))))).)))))--))))).-............. ( -43.60, z-score = -3.47, R) >droYak2.chr3L 4394041 91 + 24197627 ---------------------------UUCGCCCCUGUCCGAUCCGCCAGAU---AGCCACGGCUAUAGC---AUGGCUAUAGCUAUAGCCAAAGCUACAGCCAG--GGCGAU-GGAGAUGAGGAUG ---------------------------.........((((.(((..(((...---.(((..((((.((((---.(((((((....)))))))..)))).))))..--)))..)-)).)))..)))). ( -36.70, z-score = -2.76, R) >droAna3.scaffold_13337 11099341 121 - 23293914 AUCGCCCACCAUCACCUCCCACCAAUCCACCCGAUCUUCCAAAGCGAUGGAUAUAGCCCCCACC--AUGAGAAUGAG-GAUGGCAAUGGCCAUAGCGUU-GGCUAU-GACGAU-GGCGAUGUCGAUG (((((.(.(((((........((((((((..((...........)).)))))...(((..(..(--((....)))..-)..)))..))).((((((...-.)))))-)..)))-)).)..).)))). ( -31.40, z-score = -0.01, R) >dp4.chrXR_group8 8173019 85 - 9212921 ------------------------------------------UGCGCUGUGUUCUGUCCUCUCCUGAUCUGCUGGAGCCAUGGCAAUGGCAAUGGCAUUGGGCGAUGGGCGAUGGGCAAUGGCGAUG ------------------------------------------..(((((((((((((((((.((..(..(((((..(((((....)))))..))))))..)).)).)))))..)))).))))))... ( -34.40, z-score = -1.33, R) >consensus ___________________________UUCGCCCCGUUCCGAUCCGCCAGAU___AGCCACGGCUAUAGCUAUCUAGCCAUGGCUAUAGCCAAAGCUAUGGCCGU__GGCGAU_GGAGAUGAGGAUG .....................................(((.((.((((.(........).(((((.((((..........((((....))))..)))).)))))...)))))).))).......... (-12.56 = -13.27 + 0.71)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:45 2011