| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,740,987 – 20,741,078 |
| Length | 91 |
| Max. P | 0.795519 |

| Location | 20,740,987 – 20,741,078 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 71.80 |
| Shannon entropy | 0.53967 |
| G+C content | 0.53685 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.99 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
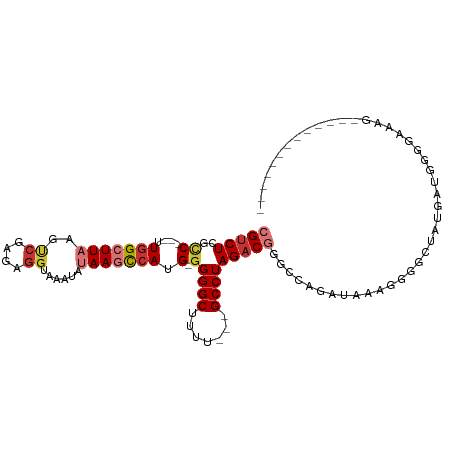
>dm3.chr3L 20740987 91 + 24543557 CGUCUCGCC---UUGGCUUAAGUCGAGAGGUAAAUAUAAGCCAUG-GGGGCUUUU---GCCUAGACGGGCCAUAUAAAGGGGCUAUGAUGGGGAAAGG------------ ((((..(((---((((((...(((...(((((((....((((...-..)))))))---)))).))).))))).......))))...))))........------------ ( -30.81, z-score = -1.57, R) >droAna3.scaffold_13337 11096315 103 + 23293914 CGUCUCGCC---UUGGCUUAAGCCGAGAGGUAAAUAUAAGCCAUG-GGGGCAUUU---GCCUAGACGGGCCUCGAAUUGUGGGGCAAAUGGCAUGAGCCGAGGGAGAGUG ..((((.((---((((((((.(((....)))........(((((.-.((((....---))))......((((((.....))))))..))))).))))))))))))))... ( -48.40, z-score = -4.53, R) >droEre2.scaffold_4784 20433629 90 + 25762168 CGUCUCGCC---UUGGCUUAAGUCGAGAGGUAAAUAUAAGCCAUG-GGGGCUUUU---GCCUAGACGAGCCCGAUAAAGGGGAUAUGAUGGAGAGAG------------- ((((((.((---(((((((..(((...(((((((....((((...-..)))))))---)))).)))))))).....)))).))...)))).......------------- ( -29.60, z-score = -1.68, R) >droYak2.chr3L 4391408 90 - 24197627 CGUCUCGGC---UUGGCUUAAGUCGAGAGGUAAAUAUAAGCCAUG-GGGGCUUUU---GCCUAGACGGGCCAGAUAAAGGGGCUACGAUGGGAAAAG------------- .(((((.(.---((((((...(((...(((((((....((((...-..)))))))---)))).))).)))))).)...)))))..............------------- ( -29.50, z-score = -1.63, R) >droSec1.super_11 630014 91 + 2888827 CGUCUCGCC---UUGGCUUCAGUCGAGAGGUAAAUAUAAGCCAUG-GGGGCUUUU---GCCUAGACGGGCCAGAUAAAGGGGCUAUGAUGGGGAAAGG------------ ((((..(((---((((((...(((...(((((((....((((...-..)))))))---)))).))).))))).......))))...))))........------------ ( -30.81, z-score = -1.19, R) >droSim1.chr3L 20097173 91 + 22553184 CGUCUCGCC---UUGGCUUAAGUCGAGAGGUAAAUAUAAGCCAUG-GGGGCUUAU---GCCUAGACGGGCCAGAUAAAGGGGCUAUGAUGGGGAAAGC------------ ((((..(((---((((((...(((...((((....(((((((...-..)))))))---)))).))).))))).......))))...))))........------------ ( -31.31, z-score = -1.77, R) >droWil1.scaffold_180949 3894571 81 - 6375548 CGGCUCGUC---CUUGGUUGAGGCGAGGUAAAUACAAAAAUCAUGCGGGGCAUUU---GCCUAGACUGAACCCGGCAACACACAGUC----------------------- (((((((.(---(((....)))))))).............(((..(.((((....---)))).)..)))..))).............----------------------- ( -20.50, z-score = 0.17, R) >dp4.chrXR_group8 8170596 94 + 9212921 CGUCUCGCCGGCUUGGCUUAAGCCGAGAGGUAAAUAUAAACCAUG-GGGGCCUUCUGGGCCUAGACUCUGCUC-UGCUCUAGUCUGGUCGGGGGUC-------------- .(.(((.((((((.((((..(((.(((.(((........)))...-.(((((.....)))))........)))-.)))..)))).))))))))).)-------------- ( -36.90, z-score = -1.21, R) >consensus CGUCUCGCC___UUGGCUUAAGUCGAGAGGUAAAUAUAAGCCAUG_GGGGCUUUU___GCCUAGACGGGCCAGAUAAAGGGGCUAUGAUGGGGAAAG_____________ (((((........(((((((..((....))......)))))))....((((.......)))))))))........................................... (-12.86 = -13.85 + 0.99)

| Location | 20,740,987 – 20,741,078 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.80 |
| Shannon entropy | 0.53967 |
| G+C content | 0.53685 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.79 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20740987 91 - 24543557 ------------CCUUUCCCCAUCAUAGCCCCUUUAUAUGGCCCGUCUAGGC---AAAAGCCCC-CAUGGCUUAUAUUUACCUCUCGACUUAAGCCAA---GGCGAGACG ------------.......((((.((((.....))))))))..(((((.(((---....)))((-(.(((((((..((........))..))))))).---)).)))))) ( -21.90, z-score = -2.00, R) >droAna3.scaffold_13337 11096315 103 - 23293914 CACUCUCCCUCGGCUCAUGCCAUUUGCCCCACAAUUCGAGGCCCGUCUAGGC---AAAUGCCCC-CAUGGCUUAUAUUUACCUCUCGGCUUAAGCCAA---GGCGAGACG ...(((((((.((((...(((....(((.(.......).)))((((...(((---....)))..-.))))................)))...)))).)---)).)))).. ( -31.50, z-score = -2.31, R) >droEre2.scaffold_4784 20433629 90 - 25762168 -------------CUCUCUCCAUCAUAUCCCCUUUAUCGGGCUCGUCUAGGC---AAAAGCCCC-CAUGGCUUAUAUUUACCUCUCGACUUAAGCCAA---GGCGAGACG -------------................(((......)))(((((((.(((---....)))..-..(((((((..((........))..))))))))---))))))... ( -25.20, z-score = -2.93, R) >droYak2.chr3L 4391408 90 + 24197627 -------------CUUUUCCCAUCGUAGCCCCUUUAUCUGGCCCGUCUAGGC---AAAAGCCCC-CAUGGCUUAUAUUUACCUCUCGACUUAAGCCAA---GCCGAGACG -------------..............(((.........))).(((((.(((---....(....-).(((((((..((........))..))))))).---))).))))) ( -19.50, z-score = -1.45, R) >droSec1.super_11 630014 91 - 2888827 ------------CCUUUCCCCAUCAUAGCCCCUUUAUCUGGCCCGUCUAGGC---AAAAGCCCC-CAUGGCUUAUAUUUACCUCUCGACUGAAGCCAA---GGCGAGACG ------------...............(((.........))).(((((.(((---..(((((..-...))))).........((......)).))).)---))))..... ( -19.70, z-score = -0.76, R) >droSim1.chr3L 20097173 91 - 22553184 ------------GCUUUCCCCAUCAUAGCCCCUUUAUCUGGCCCGUCUAGGC---AUAAGCCCC-CAUGGCUUAUAUUUACCUCUCGACUUAAGCCAA---GGCGAGACG ------------...............(((.........))).(((((.(((---....)))((-(.(((((((..((........))..))))))).---)).)))))) ( -21.70, z-score = -1.53, R) >droWil1.scaffold_180949 3894571 81 + 6375548 -----------------------GACUGUGUGUUGCCGGGUUCAGUCUAGGC---AAAUGCCCCGCAUGAUUUUUGUAUUUACCUCGCCUCAACCAAG---GACGAGCCG -----------------------..........(((.((((...((....))---....)))).)))................(((((((......))---).))))... ( -17.80, z-score = 0.65, R) >dp4.chrXR_group8 8170596 94 - 9212921 --------------GACCCCCGACCAGACUAGAGCA-GAGCAGAGUCUAGGCCCAGAAGGCCCC-CAUGGUUUAUAUUUACCUCUCGGCUUAAGCCAAGCCGGCGAGACG --------------.......(((((((((...((.-..))..))))).((((.....))))..-...)))).........((((((((((.....))))))).)))... ( -27.80, z-score = -1.69, R) >consensus _____________CUUUCCCCAUCAUAGCCCCUUUAUCUGGCCCGUCUAGGC___AAAAGCCCC_CAUGGCUUAUAUUUACCUCUCGACUUAAGCCAA___GGCGAGACG ............................................((((.(((.......))).....(((((((................)))))))........)))). (-11.13 = -11.79 + 0.66)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:44 2011