
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,728,545 – 20,728,642 |
| Length | 97 |
| Max. P | 0.866942 |

| Location | 20,728,545 – 20,728,642 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.00 |
| Shannon entropy | 0.61045 |
| G+C content | 0.46361 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -9.89 |
| Energy contribution | -9.30 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20728545 97 + 24543557 ------------UUUGGAGCGUC---AGCAC--ACUUCGCAGGUUACUGAACCUGCAAUACGCGUACUCUCGUUCAUAUGAAGCUC--GUCCAAAGCAAACAAAUUCCUAGAAAUU ------------((((((((...---.))..--.(((((((((((....))))))).....(((......)))......))))...--.))))))..................... ( -23.00, z-score = -1.63, R) >droSim1.chr3L 20085469 108 + 22553184 -GGGGUACUCUCUUUGGAGCGUC---AGCAC--ACUUCGCAGGUUACUGAACCUGCAAUACUCGUACUCUCGUUCAUAUGAAGCUC--GUCCAAAGCAAACAAAUUCCCAGAAAUU -((((......(((((((.((..---(((..--.....(((((((....))))))).............((((....)))).))))--)))))))).........))))....... ( -29.16, z-score = -2.59, R) >droSec1.super_11 616536 108 + 2888827 -GGGGUACUCUCUUUGGAGCGUC---AGCAC--ACUUCGCAGGUUACUGAACCUGCAAUACUCGUACUCUCGUUCAUAUGAAGCUC--GUCCAAAGCAAACAAAUUCCCAGAAAUU -((((......(((((((.((..---(((..--.....(((((((....))))))).............((((....)))).))))--)))))))).........))))....... ( -29.16, z-score = -2.59, R) >droYak2.chr3L 4378892 108 - 24197627 -GGGGAACUCUCUUUGGAGCGUC---AGCAC--ACUUCGCAGGUUACUGAACCUGCAAUACUCGUACUCUCGUUCGUAUGAAGCUC--GUCCAAAGCAAACAAAUUCCCAGAAAUU -.(((((....(((((((.((..---(((..--.....(((((((....))))))).....((((((........)))))).))))--))))))))........)))))....... ( -34.10, z-score = -3.64, R) >droEre2.scaffold_4784 20421261 108 + 25762168 -GGGGAACUCUCUUUGGAGCGUC---AGCAC--UCUUUGCAGGUUACUGAACCUGCAAUACUCGUACUCUGGUUCGGAUGAAGCUC--GUCCAAAGCAAACAAAUUCCCAGAAAUU -.(((((....(((((((.((..---(((..--...(((((((((....)))))))))((..((.((....)).))..))..))))--))))))))........)))))....... ( -32.70, z-score = -2.17, R) >droAna3.scaffold_13337 11081979 111 + 23293914 GGGGGAACUCUCCAUGUAGCGUC---AGCGCGUACUCAAUAGGUUACUGAACCUCCG-UGCAUACACCCU-ACCUACAUAACUCUUUAAAAAAAAGCAAACAAAUUCUAAGAAAUU .((((....))))((((((.((.---((.(.(((..((..(((((....)))))...-))..))).).))-))))))))..................................... ( -20.40, z-score = -0.98, R) >dp4.chrXR_group8 8157261 104 + 9212921 -----CUCUUUCUUUCGAGCACUUCAAGUGUGUGCUCAAUGUGUUGCUGAACCUGUGACGGACGA---CCAGCAGGUACCUAGCCG----GCUAGGCAGUAUAAUUCUCCGAAAUU -----.......((((((((((.........)))))).......(((((.((((((...((....---)).)))))).(((((...----.)))))))))).........)))).. ( -32.00, z-score = -1.59, R) >droPer1.super_40 348159 104 + 810552 -----UUCUUUCUUUCGAGCACUUCAAGUGUGUACUCAAUGUGUUGCUGAACCUGUGACGGACGA---CCAGCAGGUACCUAGCCG----GCUAGGCAGUACAAUUCCCCGAAAUU -----.......(((((.((((.....))))..........(((.((((.((((((...((....---)).)))))).(((((...----.))))))))))))......))))).. ( -28.10, z-score = -0.73, R) >consensus _GGGGAACUCUCUUUGGAGCGUC___AGCAC__ACUUCGCAGGUUACUGAACCUGCAAUACUCGUACUCUCGUUCAUAUGAAGCUC__GUCCAAAGCAAACAAAUUCCCAGAAAUU ..................((.......)).........(((((((....)))))))..........................(((.........)))................... ( -9.89 = -9.30 + -0.59)
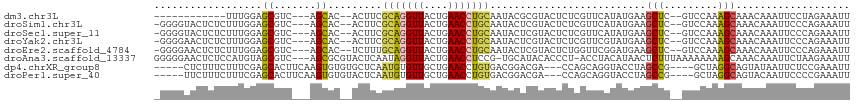

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:42 2011