| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,901,192 – 5,901,294 |
| Length | 102 |
| Max. P | 0.833289 |

| Location | 5,901,192 – 5,901,294 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.19 |
| Shannon entropy | 0.43501 |
| G+C content | 0.45082 |
| Mean single sequence MFE | -19.04 |
| Consensus MFE | -11.93 |
| Energy contribution | -12.10 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.604554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
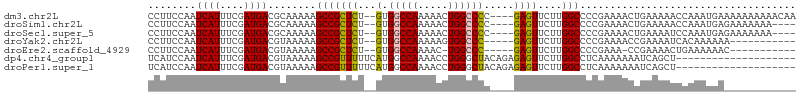
>dm3.chr2L 5901192 102 + 23011544 CCUUCCAAUCAUUUCGAUGACGCAAAAAGCCGCUCU--GUGGCCAAAAACUGGCCCC----GAGUUCUUGGCCCCGAAAACUGAAAAACCAAAUGAAAAAAAAAACAA ........((((((((.....((...(((..((((.--(.(((((.....)))))).----)))).))).))..)))))..)))........................ ( -19.50, z-score = -1.60, R) >droSim1.chr2L 5691924 98 + 22036055 CCUUCCAAUCAUUUCGAUGACGCAAAAAGCCGCUCU--GUGGCCAAAAACUGGCCCC----GAGUUCUUGGCCCCGAAAACUGAAAAACCAAAUGAGAAAAAAA---- ........((((((((.....((...(((..((((.--(.(((((.....)))))).----)))).))).))..)))))..)))....................---- ( -19.50, z-score = -1.18, R) >droSec1.super_5 3971676 98 + 5866729 CCUUCCAAUCAUUUCGAUGACGCAAAAAGCCGCUCU--GUGGCCAAAAACUGGCCCC----GAGUUCUUGGCCCCGAAAACUGAAAAUCCAAAUGAGAAAAAAA---- ........((((((.(((...((........))((.--(.((((((.(((((....)----.)))).))))))).)).........))).))))))........---- ( -21.10, z-score = -1.69, R) >droYak2.chr2L 9028343 90 - 22324452 CCUUCCAAUCAUUUCGAUGACGUAAAAAGCCGCUCU--GUGGCCAAAAAGUGGCCC-----GAGUUCUUGGCCCCGAAAACCGAAAAUCACAAAAAA----------- ...........(((((....((......(((((((.--(.(((((.....))))))-----))))....)))..)).....)))))...........----------- ( -16.90, z-score = -0.49, R) >droEre2.scaffold_4929 5986110 88 + 26641161 CCUUCCAAUCAUUUCGAUGACGUAAAAAGCCGCUCU--GUGGCCAAAAC-UGGCCC-----GAGUUCUUGGCCCCGAAA-CCGAAAACUGAAAAAAC----------- ........((((((((.(..((......(((((((.--(.(((((....-))))))-----))))....)))..))..)-.)))))..)))......----------- ( -18.50, z-score = -1.39, R) >dp4.chr4_group1 409336 88 + 5278887 UCAUCCAAUCAUUUCGAUGACGUAAAAAGCCGUUUUUCAUGGCCAAAACCUGGGCUACAGAGAGUUCUUGGCCUCAAAAAAAUCAGCU-------------------- (((((.((....)).))))).......(((.((((((...((((((((((((.....)))...))).))))))....))))))..)))-------------------- ( -18.90, z-score = -1.03, R) >droPer1.super_1 404909 88 + 10282868 UCAUCCAAUCAUUUCGAUGACGUAAAAAGCCGUUUUUCAUGGCCAAAACCUGGGCUACAGAGAGUUCUUGGCCUCAAAAAAAUCAGCU-------------------- (((((.((....)).))))).......(((.((((((...((((((((((((.....)))...))).))))))....))))))..)))-------------------- ( -18.90, z-score = -1.03, R) >consensus CCUUCCAAUCAUUUCGAUGACGUAAAAAGCCGCUCU__GUGGCCAAAAACUGGCCCC____GAGUUCUUGGCCCCGAAAACUGAAAAUCCAAAUGA____________ ...........(((((............(((((.....)))))........((((..............)))).)))))............................. (-11.93 = -12.10 + 0.16)


| Location | 5,901,192 – 5,901,294 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.19 |
| Shannon entropy | 0.43501 |
| G+C content | 0.45082 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -16.30 |
| Energy contribution | -17.43 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.833289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
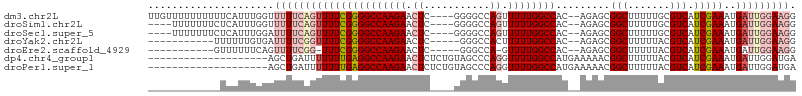
>dm3.chr2L 5901192 102 - 23011544 UUGUUUUUUUUUUCAUUUGGUUUUUCAGUUUUCGGGGCCAAGAACUC----GGGGCCAGUUUUUGGCCAC--AGAGCGGCUUUUUGCGUCAUCGAAAUGAUUGGAAGG .....(((((..((((((.(.......(((((...(((((((((((.----(....))))))))))))..--))))).((.....)).....).))))))..))))). ( -29.70, z-score = -1.69, R) >droSim1.chr2L 5691924 98 - 22036055 ----UUUUUUUCUCAUUUGGUUUUUCAGUUUUCGGGGCCAAGAACUC----GGGGCCAGUUUUUGGCCAC--AGAGCGGCUUUUUGCGUCAUCGAAAUGAUUGGAAGG ----.................(((((((((((((((((((((((((.----(....))))))))))))..--.((((((....)))).)).)))))..))))))))). ( -29.50, z-score = -1.64, R) >droSec1.super_5 3971676 98 - 5866729 ----UUUUUUUCUCAUUUGGAUUUUCAGUUUUCGGGGCCAAGAACUC----GGGGCCAGUUUUUGGCCAC--AGAGCGGCUUUUUGCGUCAUCGAAAUGAUUGGAAGG ----.(((((..((((((.(((.....(((((...(((((((((((.----(....))))))))))))..--))))).((.....))...))).))))))..))))). ( -31.60, z-score = -2.25, R) >droYak2.chr2L 9028343 90 + 22324452 -----------UUUUUUGUGAUUUUCGGUUUUCGGGGCCAAGAACUC-----GGGCCACUUUUUGGCCAC--AGAGCGGCUUUUUACGUCAUCGAAAUGAUUGGAAGG -----------..........(((((((((((((((((.(((..(((-----.(((((.....)))))..--.)))...))).....))).)))))..))))))))). ( -23.00, z-score = -0.30, R) >droEre2.scaffold_4929 5986110 88 - 26641161 -----------GUUUUUUCAGUUUUCGG-UUUCGGGGCCAAGAACUC-----GGGCCA-GUUUUGGCCAC--AGAGCGGCUUUUUACGUCAUCGAAAUGAUUGGAAGG -----------...((((((((((((((-(..(((((((.....(((-----.((((.-.....))))..--.))).)))))....))..))))))..))))))))). ( -27.90, z-score = -2.03, R) >dp4.chr4_group1 409336 88 - 5278887 --------------------AGCUGAUUUUUUUGAGGCCAAGAACUCUCUGUAGCCCAGGUUUUGGCCAUGAAAAACGGCUUUUUACGUCAUCGAAAUGAUUGGAUGA --------------------(((((.((((((...((((((((....((((.....))))))))))))..))))))))))).......(((((.((....)).))))) ( -25.30, z-score = -1.81, R) >droPer1.super_1 404909 88 - 10282868 --------------------AGCUGAUUUUUUUGAGGCCAAGAACUCUCUGUAGCCCAGGUUUUGGCCAUGAAAAACGGCUUUUUACGUCAUCGAAAUGAUUGGAUGA --------------------(((((.((((((...((((((((....((((.....))))))))))))..))))))))))).......(((((.((....)).))))) ( -25.30, z-score = -1.81, R) >consensus ____________UCAUUUGGAUUUUCAGUUUUCGGGGCCAAGAACUC____GGGGCCAGUUUUUGGCCAC__AGAGCGGCUUUUUACGUCAUCGAAAUGAUUGGAAGG .....................((((((((((((((((((((((.((......))......)))))))).........(((.......))).)))))..))))))))). (-16.30 = -17.43 + 1.13)
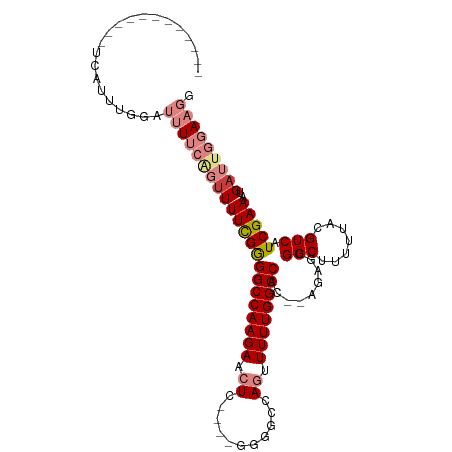

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:05 2011