
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,708,860 – 20,708,980 |
| Length | 120 |
| Max. P | 0.786474 |

| Location | 20,708,860 – 20,708,980 |
|---|---|
| Length | 120 |
| Sequences | 8 |
| Columns | 133 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Shannon entropy | 0.44649 |
| G+C content | 0.42406 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.61 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
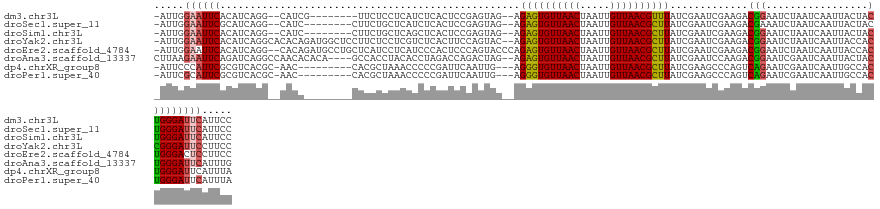
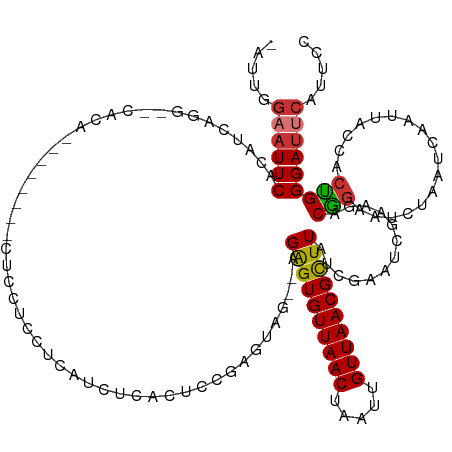
>dm3.chr3L 20708860 120 - 24543557 -AUUGGAAUUCACAUCAGG--CAUCG--------UUCUCCUCAUCUCACUCCGAGUAG--AGAGUGUUAACUAAUUGUUAACGUUUAUCGAAUCGAAGACGGAAUCUAAUCAAUUACUACUGGGAUUCAUUCC -...(((((..........--...((--------(..(((((..(((.....)))..)--))...((((((.....))))))............))..)))((((((...............))))))))))) ( -22.76, z-score = 0.47, R) >droSec1.super_11 597677 120 - 2888827 -AUUGGAAUUCGCAUCAGG--CAUC--------CUUCUGCUCAUCUCACUCCGAGUAG--AGAGUGUUAACUAAUUGUUAACGCUUAUCGAAUCGAAGACGAAAUCUAAUCAAUUACUACUGGGAUUCAUUCC -...(((((..(((..(((--...)--------))..))).........(((.(((((--.((((((((((.....))))))))))......(((....)))..............))))).)))...))))) ( -31.40, z-score = -2.44, R) >droSim1.chr3L 20063991 120 - 22553184 -AUUGGAAUUCACAUCAGG--CAUC--------CUUCUGCUCAGCUCACUCCGAGUAG--AGAGUGUUAACUAAUUGUUAACGCUUAUCGAAUCGAAGACGGAAUCUAAUCAAUUACUACUGGGAUUCAUUCC -...(((((........((--((..--------....))))........(((.(((((--.((((((((((.....))))))))))...((.((.......)).))..........))))).)))...))))) ( -29.00, z-score = -1.42, R) >droYak2.chr3L 4357325 130 + 24197627 -AUUGGAAUUCACAUCAGGCACACAGAUGGCUCCUUCUCCUCGUCUCACUUCCAGUAC--AGAGUGUUAACUAAUUGUUAACGCUUAUCGAAUCGAAGACGGAAUCUAAUCAAUUACCACCGGGAUUCCUUCC -...(((((((.((((.(.....).))))..(((.(((..(((..((((.....))..--.((((((((((.....))))))))))...))..)))))).)))...................))))))).... ( -31.70, z-score = -1.87, R) >droEre2.scaffold_4784 20401064 130 - 25762168 -AUUGGAAUUCACAUCAGG--CACAGAUGCCUGCUCAUCCUCAUCCCACUCCCAGUACCCAGAGUGUUAACUAAUUGUUAACGCUUAUCGAAUCGAAGACGGAAUCUAAUCAAUUACCACUGGGACUCCUUCC -...(((........((((--((....))))))................(((((((.....((((((((((.....))))))))))...((.((.......)).))............))))))).))).... ( -34.10, z-score = -2.93, R) >droAna3.scaffold_13337 11060579 127 - 23293914 CUUAAGAAUUCAGAUCAGGCCAACACACA----GCCACCUACACCUAGACCAGACUAG--AGAGUGUUAACUAAUUGUUAACGCUUAUCGAAUCCAAGACGGAAUCGAAUCAAUUACUACUGGGAUUCAUUUG .....(((((.......(((.........----))).............((((..(((--.((((((((((.....)))))))))).((((.(((.....))).))))........))))))))))))..... ( -32.00, z-score = -3.67, R) >dp4.chrXR_group8 8136092 119 - 9212921 -AUUCCCAUUCGCGUCACGC-AAC---------CACGCUAAACCCCCGAUUCAAUUG---AGGGUGUUAACUAAUUGUUAACGCUUAUCGAAGCCCAGUCAGAAUCGAAUCAAUUGCCACUGGGAUUCAUUUA -..(((((...((((.....-...---------.)))).............((((((---(((((((((((.....)))))))))).((((..(.......)..)))).)))))))....)))))........ ( -29.30, z-score = -2.12, R) >droPer1.super_40 327856 119 - 810552 -AUUCGCAUUCGCGUCACGC-AAC---------CACGCUAAACCCCCGAUUCAAUUG---AGGGUGUUAACUAAUUGUUAACGCUUAUCGAAGCCCAGUCAGAAUCGAAUCAAUUGCCACUGGGAUUCAUUUA -...(((....)))....((-...---------...))......((((...((((((---(((((((((((.....)))))))))).((((..(.......)..)))).)))))))....))))......... ( -28.00, z-score = -1.31, R) >consensus _AUUGGAAUUCACAUCAGG__CACA________CUCCUCCUCAUCUCACUCCGAGUAG__AGAGUGUUAACUAAUUGUUAACGCUUAUCGAAUCGAAGACGGAAUCUAAUCAAUUACCACUGGGAUUCAUUCC .....((((((..................................................((((((((((.....)))))))))).............(((.................)))))))))..... (-14.57 = -14.61 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:40 2011