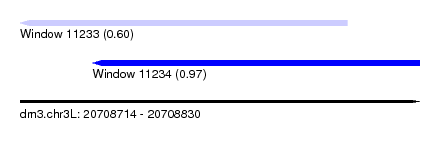
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,708,714 – 20,708,830 |
| Length | 116 |
| Max. P | 0.974732 |

| Location | 20,708,714 – 20,708,809 |
|---|---|
| Length | 95 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Shannon entropy | 0.55279 |
| G+C content | 0.53475 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -20.20 |
| Energy contribution | -19.72 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20708714 95 - 24543557 -------------------AUAUACACACCUCCAAC---UAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUGGGCGAAUCCUGCGGCGGUCCGGGCGGAUUUUCCGGUCAAUG -------------------........(((......---.((((((.(((..(((((((....((((.........)))).....))))))))))...)))))).....)))..... ( -27.40, z-score = -0.18, R) >droAna3.scaffold_13337 11060441 90 - 23293914 ------------------------GUUUUGUCCAAC---UAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUCUGGGCGAGUCCUGCGGCGGUCCGGGCGGAUUUUCCGGUCAAUG ------------------------...(((.((...---.((((((.(((..(((((((....((((.........)))).....))))))))))...)))))).....)).))).. ( -30.10, z-score = -0.53, R) >droEre2.scaffold_4784 20400923 90 - 25762168 -----------------------AUAUACCUCC-AC---UAUUCGCAGAUGCUGCCGCAUAUGCGCCAAAACCUUGGGCGAAUCCUGCGGCGGUCCGGGCGGAUUUUCCGGUCAAUG -----------------------....(((...-..---.((((((....(((((((((....((((.........)))).....)))))))))....)))))).....)))..... ( -32.20, z-score = -1.38, R) >droYak2.chr3L 4357183 91 + 24197627 -----------------------AUAUACCUCCAAC---UAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUGGGCGAAUCCUGCGGCGGCCCGGGCGGAUUUUCCGGUCAAUG -----------------------....(((......---.((((((....(((((((((....((((.........)))).....)))))))))....)))))).....)))..... ( -30.70, z-score = -0.93, R) >droSim1.chr3L 20063845 95 - 22553184 -------------------AUAUACAUACCUCCAAC---UAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUGGGCGAAUCCUGCGGCGGUCCGGGCGGAUUUUCCGGUCAAUG -------------------.....((((((......---.((((((.(((..(((((((....((((.........)))).....))))))))))...)))))).....)))..))) ( -27.40, z-score = -0.18, R) >droSec1.super_11 597531 95 - 2888827 -------------------AUAUACAUACCUCCAAC---UAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUAGGCGAAUCCUGCGGCGGUCCGGGCGGAUUUUCCGGUCAAUG -------------------.....((((((......---.((((((.(((..(((((((....((((.........)))).....))))))))))...)))))).....)))..))) ( -27.50, z-score = -0.67, R) >droVir3.scaffold_13049 6605649 100 + 25233164 -----------------UGUUUGCUCUAAUUCUAUAUUCAAUGUACAGAUGCUGUCGCAUUUGCGCCAAAACUCUCGGCGAAUCCUGCGGCGGACCCGGCGGCUUUUCCGGCCAAUG -----------------.....(((.....(((.(((.....))).)))..((((((((....((((.........)))).....))))))))....)))((((.....)))).... ( -27.20, z-score = -0.29, R) >droMoj3.scaffold_6680 12612797 117 - 24764193 AUAUUCGCAUACGUAAUCUGUGUGCUCUGUUCUGCCCUCUGCUCACAGAUGCUGUCGCAUUUGCGCCAAAACUCUGGGCGAAUCCUGCGGCGGACCCGGCGGCUUUUCCGGCCAAUG ......(((((((.....)))))))........(((.((((....))))..(((((((((((((.(((......))))))))...))))))))....)))((((.....)))).... ( -39.70, z-score = -1.69, R) >droGri2.scaffold_15110 5459444 105 + 24565398 ----------ACAUUCACAUUUACUGUAAUUCC--AUUCAAUGUUCAGAUGCUGUCGCAUUUGCGCCAAAACCUUGGGCGAAUCCUGCGGAGGACCCGGCGGCUUUUCCGGACAAUG ----------.......................--......(((((.((.(((((((..(((((.((((....)))))))))((((....))))..)))))))...)).)))))... ( -28.40, z-score = -0.72, R) >droWil1.scaffold_180949 2429162 95 - 6375548 -------------------GUAUAUAUAUGUUUAUU---CAUUUGCAGAUGUUGUCGCAUCUGCGCCAAAACUUUGGGUGAAUCCUGCGGUGGUCUGGGAGGAUUUUCCGGUCAAUG -------------------.........(((..(((---(((..((((((((....)))))))).((((....))))))))))...))).(((.(((((((...))))))))))... ( -26.00, z-score = -1.01, R) >droPer1.super_40 327719 100 - 810552 ---------------AUGUAUGUAUAUUCAUCUAUU--CCAUUCGCAGAUGUUGUCGCAUCUGCGCCAAGACCUUGGGCGAAUCCUGCGGCGGUCCCGGCGGUUUUUCCGGCCAAUG ---------------......(((.((((.......--.....(((((((((....)))))))))((((....))))..))))..)))((((....)..(((.....)))))).... ( -28.00, z-score = 0.25, R) >dp4.chrXR_group8 8135955 100 - 9212921 ---------------AUGUAUGUAUAUUCAUCUAUU--CCAUUCACAGAUGUUGUCGCAUCUGCGCCAAGACCUUGGGCGAAUCCUGCGGCGGUCCCGGCGGUUUUUCCGGCCAAUG ---------------.............(((((...--........)))))((((((((....((((.........)))).....))))))))....(((((.....)).))).... ( -24.30, z-score = 1.03, R) >anoGam1.chr3L 26214021 97 + 41284009 --------------------UCUUUCCCCACUCUCUAUUGCUGCACAGGUGCUGCAAGGUGUGUGCCCGAACGCUCGGCGAAGCUUGCGGUGGACCGGGCGGUUUUUCCGGCACGUG --------------------........((((.....((((.(((....))).)))))))).(((((.((((((((((....((.....))...)))))))....))).)))))... ( -32.70, z-score = 0.87, R) >consensus ___________________AUAUAUAUACCUCCAAC___UAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUGGGCGAAUCCUGCGGCGGUCCGGGCGGAUUUUCCGGUCAAUG ...................................................((((((((....((((.........)))).....))))))))....(((((.....)).))).... (-20.20 = -19.72 + -0.48)

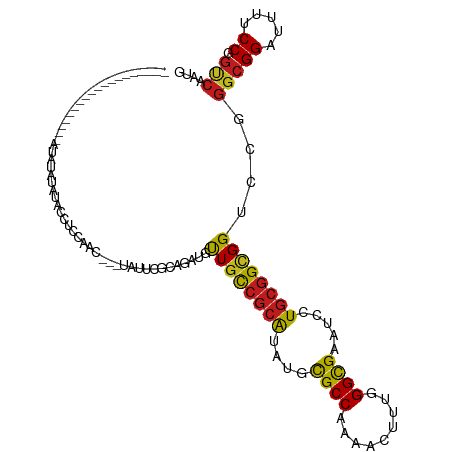
| Location | 20,708,735 – 20,708,830 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 64.27 |
| Shannon entropy | 0.74477 |
| G+C content | 0.47945 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -16.37 |
| Energy contribution | -15.88 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.51 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20708735 95 - 24543557 --------------------CAUCUAUUUAUUCUAUAUGUCAUAUACACACCUCCAACUAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUGGGCGAAUCCUGCGGCGGUCC --------------------.............................(((...(((...........)))((((((....((((.........)))).....))))))))).. ( -19.20, z-score = -0.14, R) >droAna3.scaffold_13337 11060462 84 - 23293914 -------------------------------UAUACAUAUCCGGUCGUUUUGUCCAACUAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUCUGGGCGAGUCCUGCGGCGGUCC -------------------------------...((((...(((..(((......)))...)))...))))(((((((....((((.........)))).....))))))).... ( -21.10, z-score = 0.53, R) >droEre2.scaffold_4784 20400944 90 - 25762168 --------------------CAUCCAUCUACACUAUAUGUCAUAU----ACCUCC-ACUAUUCGCAGAUGCUGCCGCAUAUGCGCCAAAACCUUGGGCGAAUCCUGCGGCGGUCC --------------------.........................----......-.............(((((((((....((((.........)))).....))))))))).. ( -22.00, z-score = -0.93, R) >droYak2.chr3L 4357204 91 + 24197627 --------------------CAUCCAUUUAUACUAUAUGUCAUAU----ACCUCCAACUAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUGGGCGAAUCCUGCGGCGGCCC --------------------.........................----....................(((((((((....((((.........)))).....))))))))).. ( -21.40, z-score = -0.65, R) >droSim1.chr3L 20063866 95 - 22553184 --------------------CAUCCAUUUAUUCUAUAUGUCAUAUACAUACCUCCAACUAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUGGGCGAAUCCUGCGGCGGUCC --------------------...((..........(((((.....)))))......................((((((....((((.........)))).....))))))))... ( -20.70, z-score = -0.50, R) >droSec1.super_11 597552 95 - 2888827 --------------------CAUCCAUUUAUUCUAUAUGUCAUAUACAUACCUCCAACUAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUAGGCGAAUCCUGCGGCGGUCC --------------------...((..........(((((.....)))))......................((((((....((((.........)))).....))))))))... ( -20.80, z-score = -1.14, R) >droMoj3.scaffold_6680 12612818 115 - 24764193 AUUCAUUUAAUGUGCAUCGAUAUUCGCAUACGUAAUCUGUGUGCUCUGUUCUGCCCUCUGCUCACAGAUGCUGUCGCAUUUGCGCCAAAACUCUGGGCGAAUCCUGCGGCGGACC ..........(((((..........)))))....(((((((.((...............)).))))))).(((((((((((((.(((......))))))))...))))))))... ( -33.96, z-score = -1.55, R) >droGri2.scaffold_15110 5459465 105 + 24565398 -------CAUAAAUUUGAUGUGUGCUUCACAUUCACAUUUACUGUAAUUCCAUUCAA-UGUUC--AGAUGCUGUCGCAUUUGCGCCAAAACCUUGGGCGAAUCCUGCGGAGGACC -------..(((((.(((((((.....)))).))).)))))(((((...........-....(--((((((....)))))))((((.........)))).....)))))...... ( -23.60, z-score = -0.07, R) >droWil1.scaffold_180949 2429183 94 - 6375548 ---------------------AUACACUUGGUAUAACGUUUGUAUAUAUAUGUUUAUUCAUUUGCAGAUGUUGUCGCAUCUGCGCCAAAACUUUGGGUGAAUCCUGCGGUGGUCU ---------------------(((((..((......))..))))).....(((..((((((..((((((((....)))))))).((((....))))))))))...)))....... ( -23.10, z-score = -0.93, R) >droPer1.super_40 327740 99 - 810552 ---------------CCUAAUAUGCACUCUAAUUUAUGUAUGUAUAUUCAUCUAU-UCCAUUCGCAGAUGUUGUCGCAUCUGCGCCAAGACCUUGGGCGAAUCCUGCGGCGGUCC ---------------...((((((((..(........)..)))))))).......-.((....((((((((....))))))))(((....(....)(((.....))))))))... ( -25.50, z-score = -1.21, R) >dp4.chrXR_group8 8135976 99 - 9212921 ---------------CCUAAUAUGCACUCUAAUUUAUGUAUGUAUAUUCAUCUAU-UCCAUUCACAGAUGUUGUCGCAUCUGCGCCAAGACCUUGGGCGAAUCCUGCGGCGGUCC ---------------...((((((((..(........)..))))))))(((((..-.........)))))((((((((....((((.........)))).....))))))))... ( -23.10, z-score = -0.80, R) >anoGam1.chr3L 26214042 90 + 41284009 -------------------------UUGCUCUGCAUUUCUCUUUCCCCACUCUCUAUUGCUGCACAGGUGCUGCAAGGUGUGUGCCCGAACGCUCGGCGAAGCUUGCGGUGGACC -------------------------...(.(((((((((........(((.(.((.((((.(((....))).)))))).).))).((((....)))).))))..))))).).... ( -23.40, z-score = 1.28, R) >consensus ____________________CAUCCAUUUAUUCUAUAUGUCAUAUACAUACCUCCAACUAUUCGCAGAUGUUGCCGCAUAUGCGCCAAAACUUUGGGCGAAUCCUGCGGCGGUCC ......................................................................((((((((....((((.........)))).....))))))))... (-16.37 = -15.88 + -0.48)
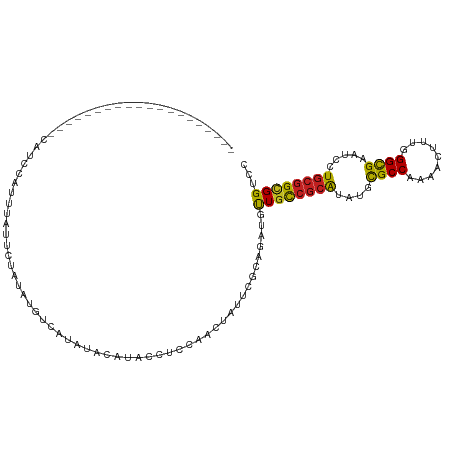
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:39 2011