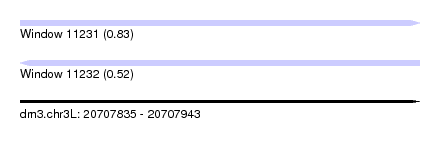
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,707,835 – 20,707,943 |
| Length | 108 |
| Max. P | 0.829521 |

| Location | 20,707,835 – 20,707,943 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 71.24 |
| Shannon entropy | 0.54843 |
| G+C content | 0.36487 |
| Mean single sequence MFE | -19.99 |
| Consensus MFE | -8.28 |
| Energy contribution | -8.41 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
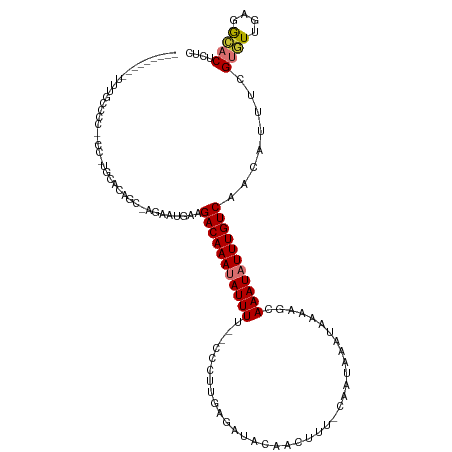
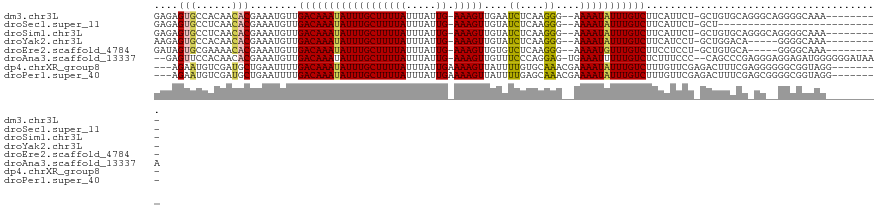
>dm3.chr3L 20707835 108 + 24543557 ---------UUUGCCCCUGCCCUGCACAGC-AGAAUGAAGACAAAUAUUUU--CCCUUGAGAUUCAACUUU-CAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGUGGCACUCUC ---------..((((...((...))(((((-((((((..(((((((((((.--...((((((......)))-)))...........)))))))))))..)))))..))))))))))..... ( -24.36, z-score = -2.58, R) >droSec1.super_11 596649 90 + 2888827 ---------------------------AGC-AGAAUGAAGACAAAUAUUUU--CCCUUGAGAUACAACUUU-CAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGAGGCACUCUC ---------------------------...-.(((((..(((((((((((.--...((((((......)))-)))...........)))))))))))..))))).((((....)))).... ( -16.76, z-score = -1.57, R) >droSim1.chr3L 20062959 108 + 22553184 ---------UUUGCCCCUGCCCUGCACAGC-AGAAUGAAGACAAAUAUUUU--CCCUUGAGAUACAACUUU-CAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGAGGCACUCUC ---------..((((..(((...)))((((-((((((..(((((((((((.--...((((((......)))-)))...........)))))))))))..)))))..))))).))))..... ( -23.96, z-score = -2.66, R) >droYak2.chr3L 4356307 103 - 24197627 ---------UUUGCCCC-----UGUCCAGC-AGGAUGAAGACAAAUAUUUU--CCCUUGAGAUACAACUUU-CAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGUGGCACUCUU ---------........-----((((((((-(.((((..(((((((((((.--...((((((......)))-)))...........)))))))))))..))..)).))))).))))..... ( -22.86, z-score = -2.13, R) >droEre2.scaffold_4784 20400120 103 + 25762168 ---------UUUGCCCC-----UGCACAGC-AGGAGGAAGACAAACAUUUU--CCCUUGAGACACAACUUU-CAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUUUCGCACUAUC ---------(((((.((-----(((...))-))).((((((......))))--)).((((((......)))-))).........)))))................((((....)))).... ( -19.30, z-score = -1.53, R) >droAna3.scaffold_13337 11059528 115 + 23293914 UUUAUCCCCCCAUCUCCUCCCUCGGGCUG--GGGAAAGAGACAAAAAUUUCA-CUCCUGGGAAACAACUUU-CAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGUGGAACUC-- ....(((.(((((((((.((....))..)--))))..((((......)))).-....))))..(((((...-..(((((((........)))))))..((.....))))))))))....-- ( -22.40, z-score = 0.40, R) >dp4.chrXR_group8 8135254 110 + 9212921 --------CCUACCGCCCCCCUCGAAAGUCUCGAACAAAGACAAAUAUUUUCGUUUGCACAAAAUAACUUUUCAAUAAAUAAAAGCAAAUAUUUGUCAAAAUUCAGCAUCGACAUUCU--- --------......((.....((((.....)))).....(((((((((((..((((.....))))..(((((........))))).)))))))))))........))...........--- ( -14.90, z-score = -2.70, R) >droPer1.super_40 327027 110 + 810552 --------CCUACCGCCCCGCUCGAAAGUCUCGAACAAAGACAAAUAUUUUCGUUUGCUCAAAAUAACUUUUCAAUAAAUAAAAGCAAAUAUUUGUCAAAAUUCAGCAUCGACAUUCU--- --------......((...(.((((.....)))).)...(((((((((((..((((.....))))..(((((........))))).)))))))))))........))...........--- ( -15.40, z-score = -2.08, R) >consensus _________UUUGCCCC__CC_UGCACAGC_AGAAUGAAGACAAAUAUUUU__CCCUUGAGAUACAACUUU_CAAUAAAUAAAAGCAAAUAUUUGUCAACAUUUCGUGUUGAGGCACUCUC .......................................(((((((((((....................................)))))))))))........((((....)))).... ( -8.28 = -8.41 + 0.12)


| Location | 20,707,835 – 20,707,943 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 71.24 |
| Shannon entropy | 0.54843 |
| G+C content | 0.36487 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.05 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20707835 108 - 24543557 GAGAGUGCCACAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUG-AAAGUUGAAUCUCAAGGG--AAAAUAUUUGUCUUCAUUCU-GCUGUGCAGGGCAGGGGCAAA--------- .....((((.........((((..((((((((((((((((((.....))-)))))....(((....))--))))))))))))..))))((-(((......))))).))))..--------- ( -30.70, z-score = -2.38, R) >droSec1.super_11 596649 90 - 2888827 GAGAGUGCCUCAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUG-AAAGUUGUAUCUCAAGGG--AAAAUAUUUGUCUUCAUUCU-GCU--------------------------- .((((((....((((.....))))((((((((((((((((((.....))-)))))....(((....))--))))))))))))..))))))-...--------------------------- ( -21.40, z-score = -2.32, R) >droSim1.chr3L 20062959 108 - 22553184 GAGAGUGCCUCAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUG-AAAGUUGUAUCUCAAGGG--AAAAUAUUUGUCUUCAUUCU-GCUGUGCAGGGCAGGGGCAAA--------- .....((((((.......((((..((((((((((((((((((.....))-)))))....(((....))--))))))))))))..))))((-((...))))....))))))..--------- ( -31.80, z-score = -2.67, R) >droYak2.chr3L 4356307 103 + 24197627 AAGAGUGCCACAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUG-AAAGUUGUAUCUCAAGGG--AAAAUAUUUGUCUUCAUCCU-GCUGGACA-----GGGGCAAA--------- .....((((..........(((..((((((((((((((((((.....))-)))))....(((....))--))))))))))))..)))(((-(.....))-----))))))..--------- ( -28.20, z-score = -2.53, R) >droEre2.scaffold_4784 20400120 103 - 25762168 GAUAGUGCGAAAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUG-AAAGUUGUGUCUCAAGGG--AAAAUGUUUGUCUUCCUCCU-GCUGUGCA-----GGGGCAAA--------- ....(((......)))........((((((((((((((((((.....))-)))))....(((....))--))))))))))))...(((((-((...)))-----))))....--------- ( -27.40, z-score = -2.06, R) >droAna3.scaffold_13337 11059528 115 - 23293914 --GAGUUCCACAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUG-AAAGUUGUUUCCCAGGAG-UGAAAUUUUUGUCUCUUUCCC--CAGCCCGAGGGAGGAGAUGGGGGGAUAAA --..(((((....(((.(((((((....)))))))(((((((.....))-)))))............)-)).....((..(((((((((.--(.....).)))))))))..)))))))... ( -29.70, z-score = -0.28, R) >dp4.chrXR_group8 8135254 110 - 9212921 ---AGAAUGUCGAUGCUGAAUUUUGACAAAUAUUUGCUUUUAUUUAUUGAAAAGUUAUUUUGUGCAAACGAAAAUAUUUGUCUUUGUUCGAGACUUUCGAGGGGGGCGGUAGG-------- ---....(.((((..((((((...(((((((((((((((((........))))))....((((....)))))))))))))))...)))).))....)))).)...........-------- ( -23.10, z-score = -0.81, R) >droPer1.super_40 327027 110 - 810552 ---AGAAUGUCGAUGCUGAAUUUUGACAAAUAUUUGCUUUUAUUUAUUGAAAAGUUAUUUUGAGCAAACGAAAAUAUUUGUCUUUGUUCGAGACUUUCGAGCGGGGCGGUAGG-------- ---..........(((((......(((((((((((((((......(((....)))......)))))......))))))))))((((((((((...)))))))))).)))))..-------- ( -26.00, z-score = -1.45, R) >consensus GAGAGUGCCACAACACGAAAUGUUGACAAAUAUUUGCUUUUAUUUAUUG_AAAGUUGUAUCUCAAGGG__AAAAUAUUUGUCUUCAUUCU_GCUGUGCA_GG__GGGGCAAA_________ ....(((......)))........((((((((((((((((((.....)).)))))....((....))....)))))))))))....................................... ( -9.56 = -9.05 + -0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:37 2011