| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,882,320 – 5,882,458 |
| Length | 138 |
| Max. P | 0.899525 |

| Location | 5,882,320 – 5,882,458 |
|---|---|
| Length | 138 |
| Sequences | 6 |
| Columns | 153 |
| Reading direction | reverse |
| Mean pairwise identity | 80.55 |
| Shannon entropy | 0.35655 |
| G+C content | 0.62477 |
| Mean single sequence MFE | -49.68 |
| Consensus MFE | -31.93 |
| Energy contribution | -34.60 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
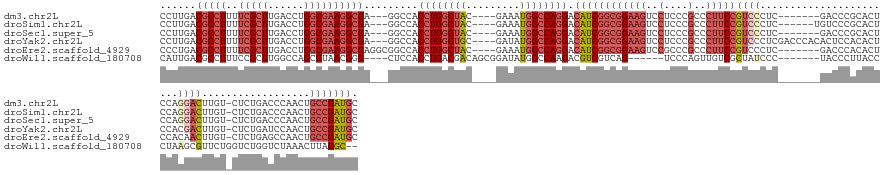
>dm3.chr2L 5882320 138 - 23011544 CCUUGACGCCUUUUCGCUUGACCUGGCGAAGGCGA---GGCCACCUGGCUAC----GAAAUGGCCAGGACAUCGGCGGAAGUCCUCCCGCCCUUCCGUCCCUC-------GACCCGCACUCCAGGACUUGU-CUCUGACCCAACUGCCGAUGC (((...(((((..(((((......)))))))))))---))...((((((((.----....)))))))).((((((((((((((((...((......(((....-------)))..)).....)))))))((-(...)))....))))))))). ( -53.00, z-score = -2.72, R) >droSim1.chr2L 5672872 139 - 22036055 CCUUGACGCCUUUUCGCUUGACCUGGCGAAGGCGA---GGCCACCUGGCUAC----GAAAUGGCCAGGACAUCGGCGGAAGUCCUCCCGCCCUUCCGUCCCUC------UGUCCCGCACUCCAGGACUUGU-CUCUGACCCAACUGCCGAUGC (((...(((((..(((((......)))))))))))---))...((((((((.----....)))))))).((((((((((((..(....)..)))))(((....------.((((.(.....).))))....-....)))......))))))). ( -52.52, z-score = -2.49, R) >droSec1.super_5 3952150 138 - 5866729 CCUUGACGCCUUUUCGCUUGACCUGGCGAAGGCGA---GGCCACCUGGCUAC----GAAAUGGCCAGGACAUCGGCGGAAGUCCUCCCGCCCUUCCGUCCCUC-------GACCCGCACUCCAGGACUUGU-CUCUGACCCAACUGCCGAUGC (((...(((((..(((((......)))))))))))---))...((((((((.----....)))))))).((((((((((((((((...((......(((....-------)))..)).....)))))))((-(...)))....))))))))). ( -53.00, z-score = -2.72, R) >droYak2.chr2L 9009264 145 + 22324452 CCUUGACGCCUUUUCGCUUGACCUGGCGAAGGCGA---GGCCACCUGGCUGC----GAUAUGGCCAGGACAUCGGCGGAAGUCCUCCCGCCCUUCCGUCCCUCGACCCACACUCCACACUCCACGACUUGU-CUCUGAUCCAACUGCCGAUGC (((...(((((..(((((......)))))))))))---))...(((((((..----.....))))))).((((((((((((..(....)..)))))(((....))).........................-.............))))))). ( -48.10, z-score = -1.81, R) >droEre2.scaffold_4929 5966081 141 - 26641161 CCCUGACGCCUUUUCGCUUGACCUGGCGAAGGCGAGGCGGCCACCUGGCUAC----GAAAUGGCCAGGACAUCGGCGGAAGUCCGCCCGCCCUUCCGUCCCUC-------GACCCACACUCCACAACUUGU-CUCUGAGCCAACUGCCGAUGC ......(((((..(((((......)))))))))).(((((((....))))..----....(((((((((((..(((((....))))).........(((....-------)))...............)))-).))).))))...)))..... ( -51.90, z-score = -1.99, R) >droWil1.scaffold_180708 4410558 134 + 12563649 CAUUGACGCCUUUCCGCUUGGCCAGCCUAACGGG----CUCCACCUGACGACAGCGGAUAUGGCCAAGACGUCGUCAG------UCCCAGUUGUCGCUAUCCC-------UACCCUUACCCUAAGCGUUCUGGUCUGGUCUAAACUUAGGC-- .......((((...(((((((((((((.....))----)(((..(((....))).)))..)))))))).))...((((------..((((....((((.....-------.............))))..)))).)))).........))))-- ( -39.57, z-score = -0.44, R) >consensus CCUUGACGCCUUUUCGCUUGACCUGGCGAAGGCGA___GGCCACCUGGCUAC____GAAAUGGCCAGGACAUCGGCGGAAGUCCUCCCGCCCUUCCGUCCCUC_______GACCCGCACUCCAGGACUUGU_CUCUGACCCAACUGCCGAUGC ......(((((..(((((......)))))))))).........((((((((.........)))))))).((((((((((((..(....)..)))))((((.......................))))..................))))))). (-31.93 = -34.60 + 2.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:04 2011