| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,642,711 – 20,642,781 |
| Length | 70 |
| Max. P | 0.701928 |

| Location | 20,642,711 – 20,642,781 |
|---|---|
| Length | 70 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 85.07 |
| Shannon entropy | 0.27669 |
| G+C content | 0.53347 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -16.28 |
| Energy contribution | -18.17 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
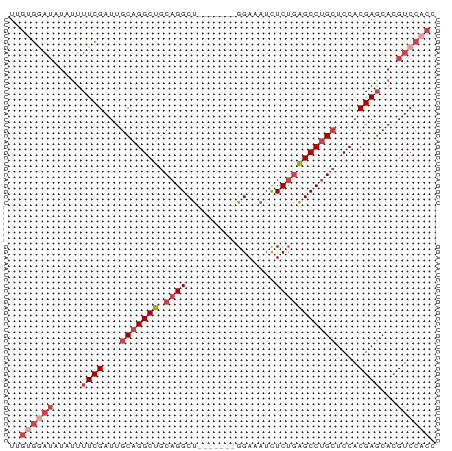
>dm3.chr3L 20642711 70 + 24543557 UUGUGGAUAUAUUUUCGAUUGCAGGCUGCAGGCU-------GGAAAUCUCUGAGCCUGCUCCACGAGCACGUCCACC ..((((((.....((((...(((((((.((((.(-------....).).))))))))))....))))...)))))). ( -25.80, z-score = -2.56, R) >droSim1.chr3L 19995033 70 + 22553184 UUGUGGAUAUAUUUUCGAUUGCAGGCUGCAGGCU-------GGAAAUCUCUGAGCCUGCUCCACGAGCACGUCCACC ..((((((.....((((...(((((((.((((.(-------....).).))))))))))....))))...)))))). ( -25.80, z-score = -2.56, R) >droSec1.super_11 532020 70 + 2888827 UUGUGGAUAUAUUUUCGAUUGCAGGCUGCAGGCU-------GGAAAUCUCUGAGCCUGCUCCACGAGCACGUCCACC ..((((((.....((((...(((((((.((((.(-------....).).))))))))))....))))...)))))). ( -25.80, z-score = -2.56, R) >droYak2.chr3L 4289942 69 - 24197627 UUGUGAAUAU-UUUUCGAUUGCAGGCUGCAGGCU-------GGAUAUCUCUGAGCCUGCUCCACGAGCACGUCCACC ..(((.((..-..((((...(((((((.((((..-------.......)))))))))))....))))...)).))). ( -19.20, z-score = -0.68, R) >droEre2.scaffold_4784 20336032 70 + 25762168 UUGCGGAUAAAUUUUCGAUUGCAGGCUGCAGGCU-------GGAAAUCUCGGAGCCUGCUCCACGAGCACGUCCACC ....((((.....((((...(((((((.(..(.(-------....).)..).)))))))....))))...))))... ( -22.50, z-score = -1.01, R) >droAna3.scaffold_13337 10994604 69 + 23293914 --------UUCAUUUCGAUUGCAGGCCACAGGCCUGCCAUAGGAAAUUUCUUGGCCAGGUCCACGAUCACGUCCACC --------............(((((((...)))))))....(((....((.(((......))).)).....)))... ( -19.60, z-score = -1.60, R) >consensus UUGUGGAUAUAUUUUCGAUUGCAGGCUGCAGGCU_______GGAAAUCUCUGAGCCUGCUCCACGAGCACGUCCACC ..((((((.....((((...(((((((.((((................)))))))))))....))))...)))))). (-16.28 = -18.17 + 1.89)


| Location | 20,642,711 – 20,642,781 |
|---|---|
| Length | 70 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 85.07 |
| Shannon entropy | 0.27669 |
| G+C content | 0.53347 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -16.90 |
| Energy contribution | -18.57 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.615189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
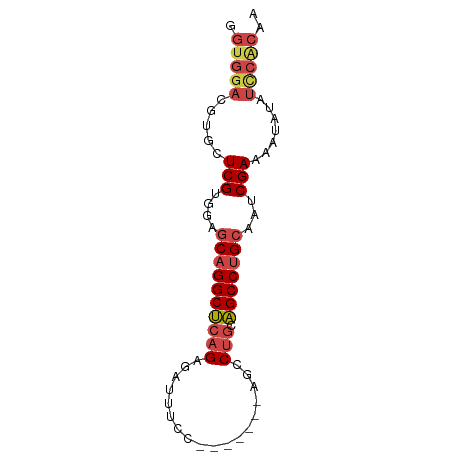
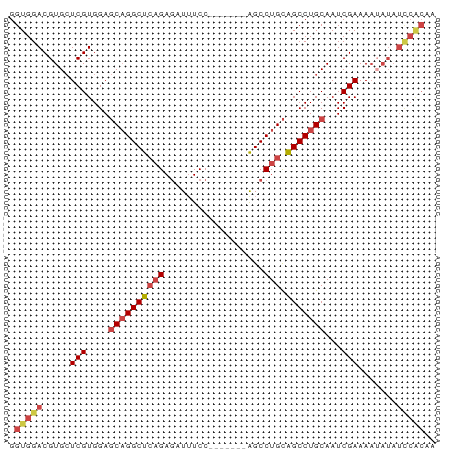
>dm3.chr3L 20642711 70 - 24543557 GGUGGACGUGCUCGUGGAGCAGGCUCAGAGAUUUCC-------AGCCUGCAGCCUGCAAUCGAAAAUAUAUCCACAA .(((((..((.(((....((((((((((.(......-------..)))).)))))))...)))...))..))))).. ( -25.70, z-score = -2.41, R) >droSim1.chr3L 19995033 70 - 22553184 GGUGGACGUGCUCGUGGAGCAGGCUCAGAGAUUUCC-------AGCCUGCAGCCUGCAAUCGAAAAUAUAUCCACAA .(((((..((.(((....((((((((((.(......-------..)))).)))))))...)))...))..))))).. ( -25.70, z-score = -2.41, R) >droSec1.super_11 532020 70 - 2888827 GGUGGACGUGCUCGUGGAGCAGGCUCAGAGAUUUCC-------AGCCUGCAGCCUGCAAUCGAAAAUAUAUCCACAA .(((((..((.(((....((((((((((.(......-------..)))).)))))))...)))...))..))))).. ( -25.70, z-score = -2.41, R) >droYak2.chr3L 4289942 69 + 24197627 GGUGGACGUGCUCGUGGAGCAGGCUCAGAGAUAUCC-------AGCCUGCAGCCUGCAAUCGAAAA-AUAUUCACAA .(((((..(..(((....((((((((((.(......-------..)))).)))))))...)))...-)..))))).. ( -23.30, z-score = -1.77, R) >droEre2.scaffold_4784 20336032 70 - 25762168 GGUGGACGUGCUCGUGGAGCAGGCUCCGAGAUUUCC-------AGCCUGCAGCCUGCAAUCGAAAAUUUAUCCGCAA .(((((.((..(((....(((((((.((.(......-------..).)).)))))))...)))..))...))))).. ( -21.10, z-score = -0.20, R) >droAna3.scaffold_13337 10994604 69 - 23293914 GGUGGACGUGAUCGUGGACCUGGCCAAGAAAUUUCCUAUGGCAGGCCUGUGGCCUGCAAUCGAAAUGAA-------- (((..(((....)))..)))..........(((((.....(((((((...)))))))....)))))...-------- ( -21.60, z-score = -0.91, R) >consensus GGUGGACGUGCUCGUGGAGCAGGCUCAGAGAUUUCC_______AGCCUGCAGCCUGCAAUCGAAAAUAUAUCCACAA .(((((.(((.(((....((((((((((..................))).)))))))...)))...))).))))).. (-16.90 = -18.57 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:27 2011