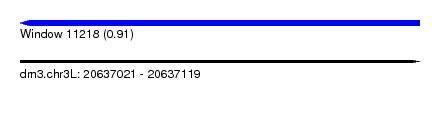
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,637,021 – 20,637,119 |
| Length | 98 |
| Max. P | 0.905341 |

| Location | 20,637,021 – 20,637,119 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.71 |
| Shannon entropy | 0.60915 |
| G+C content | 0.37350 |
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -8.80 |
| Energy contribution | -8.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
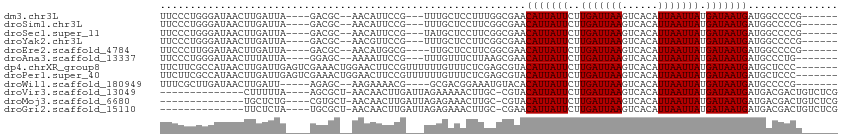
>dm3.chr3L 20637021 98 - 24543557 UUCCCUGGGAUAACUUGAUUA----GACGC--AACAUUCCG---UUUGCUCCUUUGGCGAACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGGCCCCG------ ......((........((.((----((((.--.......))---)))).))....(((.(.(((((((..(((((((......))))))).))))))).).))))).------ ( -20.70, z-score = -1.40, R) >droSim1.chr3L 19989385 98 - 22553184 UUCCCUGGGAUAACUUGAUUA----GACGC--AACAUUCCG---UUUGCUCCUUCGGCGAACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGGCCCCG------ ......((........((.((----((((.--.......))---)))).))....(((.(.(((((((..(((((((......))))))).))))))).).))))).------ ( -21.00, z-score = -1.35, R) >droSec1.super_11 526371 98 - 2888827 UUCCCUGGGAUAACUUGAUUA----GACGC--AACAUUCCG---UAUGCUCCUUCGGCGAACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGGCCCCG------ ......((........((...----((.((--(((.....)---).)))))..))(((.(.(((((((..(((((((......))))))).))))))).).))))).------ ( -20.10, z-score = -0.97, R) >droYak2.chr3L 4284186 98 + 24197627 UUCCCUGGGAUAACUUGAUUA----GACGC--AACGUUCCG---UUUGCUCCUUCGGCGAACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGGCCCCG------ ......(((...(((((((((----((((.--..))))..(---((((((.....)))))))........)))))))))(((((((.......))))).))..))).------ ( -21.40, z-score = -1.19, R) >droEre2.scaffold_4784 20330596 97 - 25762168 UUCCCUUGGAUAACUUGAUUA----GACGC--AACAUGGCG----UUGCUCCUUCGGCGAACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGGCCCCG------ .......((.......((...----((.((--(((.....)----))))))..))(((.(.(((((((..(((((((......))))))).))))))).).))))).------ ( -23.70, z-score = -2.03, R) >droAna3.scaffold_13337 10989180 97 - 23293914 UUCCCUGGGAUAACUUUAUUA----GGAGC--AAAAUUCCG---UUUGUUUCUUAAGCGAACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGCCCUG------- ......(((....(((....(----(((((--(((......---))))))))).)))....(((((((..(((((((......))))))).)))))))...)))..------- ( -20.70, z-score = -2.12, R) >dp4.chrXR_group8 8060413 106 - 9212921 UUCUUCGCCAUAACUUGAUUGAGUCGAAACUGGAACUUCCGUUUUUUGUUUCUCGAGCGUACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGCUCCC------- ............((((....)))).(((((.(((((....)))))..)))))..((((((.(((((((..(((((((......))))))).)))))))))))))..------- ( -24.80, z-score = -2.89, R) >droPer1.super_40 253573 106 - 810552 UUCUUCGCCAUAACUUGAUUGAGUCGAAACUGGAACUUCCGUUUUUUGUUUCUCGAGCGUACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGCUCCC------- ............((((....)))).(((((.(((((....)))))..)))))..((((((.(((((((..(((((((......))))))).)))))))))))))..------- ( -24.80, z-score = -2.89, R) >droWil1.scaffold_180949 2321547 95 - 6375548 UUUCGCUUGAUAACUUGAUU-----AGAGC--AAGAAAACG----GCGACGGAAAUGUACACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGCCCCG------- ..(((((......((((...-----....)--))).....)----))))(((....(((..(((((((..(((((((......))))))).))))))).))).)))------- ( -17.90, z-score = -1.40, R) >droVir3.scaffold_13049 6526979 93 + 25233164 --------------CUUUUUA----AGCGCU-AACAACUUGAUUAGAAAAACUUGC-CGUACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGACGACUGUCUCG --------------((..(((----((....-.....)))))..))......(((.-(((.(((((((..(((((((......))))))).)))))))))).)))........ ( -13.40, z-score = -0.72, R) >droMoj3.scaffold_6680 12527108 93 - 24764193 --------------UGCUCUG----CGUGCU-AACAACUUGAUUAGAGAAACUUGC-CGUACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGACGACUGUCUCG --------------.((....----...)).-.............((((...(((.-(((.(((((((..(((((((......))))))).)))))))))).)))...)))). ( -18.10, z-score = -1.30, R) >droGri2.scaffold_15110 5385817 93 + 24565398 --------------UUCUCUA----UGCGCU-AACAACUUGAUUAGAGAAACUUGC-CGAACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGACGACUGUCUCG --------------(((((((----......-...........)))))))......-(((.(((((((..(((((((......))))))).)))))))..(((....)))))) ( -16.53, z-score = -1.55, R) >consensus UUCCCUGGGAUAACUUGAUUA____GACGC__AACAUUCCG___UUUGCUCCUUGGGCGAACAUUAUUCUUGAUUAAGUCACAUUAAUUAUGAUAAUGAUGGCCCCG______ .............................................................(((((((..(((((((......))))))).)))))))............... ( -8.80 = -8.80 + -0.00)
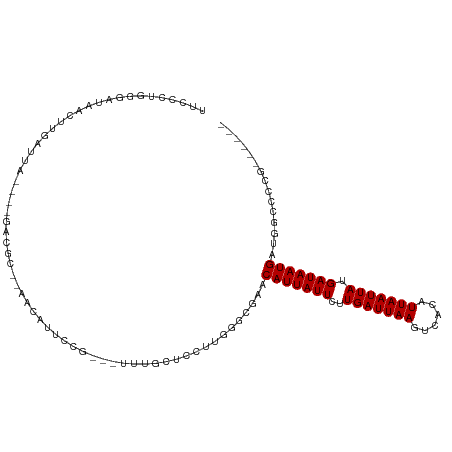
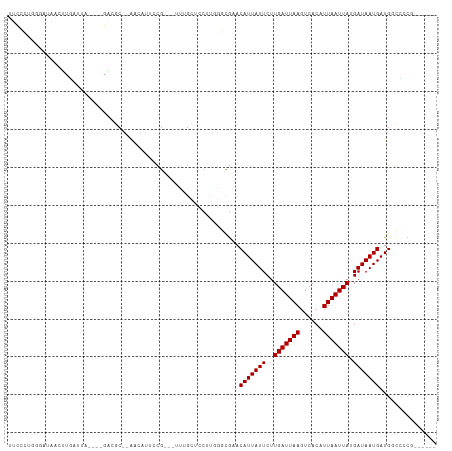
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:26 2011