
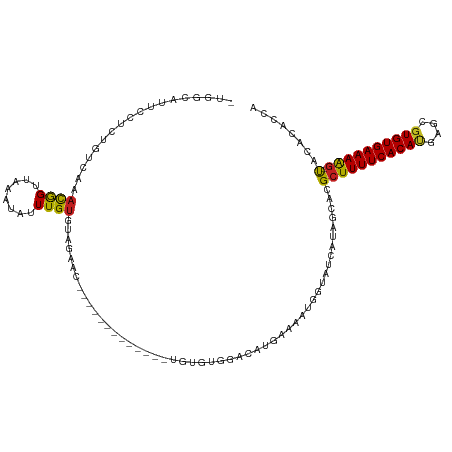
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,881,057 – 5,881,161 |
| Length | 104 |
| Max. P | 0.957691 |

| Location | 5,881,057 – 5,881,161 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.29 |
| Shannon entropy | 0.64233 |
| G+C content | 0.40537 |
| Mean single sequence MFE | -28.39 |
| Consensus MFE | -10.97 |
| Energy contribution | -10.62 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5881057 104 + 23011544 -UGGCAUUCCUCUGACCAACGGUUAAAUAUUUGUGCAGCAC-------------UGUGCGGACAUGAAAAUGGUAUCAUAGCACGCUUUUCACACAGCCGUGUGAAAAG-UACACACCA -(((((......)).)))..(((.........((((.((..-------------...))...(((....)))........))))(((((((((((....))))))))))-)....))). ( -30.30, z-score = -1.55, R) >droSim1.chr2L 5671616 104 + 22036055 -UGGCAUGCCUCUGCCCAACGGUUAAAUAUUUGUGCAGCAC-------------UGUGCGGACAUGAAAAUGGUAUCAUAGCACGCUUUUCACACAGCCGUGUGAAAAG-UACACACCG -.((((......))))...((((.........((((.((..-------------...))...(((....)))........))))(((((((((((....))))))))))-)....)))) ( -33.40, z-score = -1.86, R) >droSec1.super_5 3950893 104 + 5866729 -UGGCAUGCCUCUGCCCAACGGUUAAAUAUUUGUGCAGCAC-------------UGUGCGGACAUGAAAAUGGUAUCAUAGCACGCUUUUCACACAGCCGUGUGAAAAG-UACACACCG -.((((......))))...((((.........((((.((..-------------...))...(((....)))........))))(((((((((((....))))))))))-)....)))) ( -33.40, z-score = -1.86, R) >droYak2.chr2L 9007980 104 - 22324452 -UGGCAUUCCUCUGACCAACGGUUAAAUAUUUGUGUAGCAC-------------UGUGUGGACAUGAAAAUGGUAUCAUAGCACGCUUUUCACACAGCCGUGUGAAAAG-UACACACCG -.((........(((((...)))))......(((((....(-------------((((....(((....)))....)))))...(((((((((((....))))))))))-)))))))). ( -30.60, z-score = -1.74, R) >droEre2.scaffold_4929 5964883 103 + 26641161 -UGGCAUUCCUCUGCCCAACGGUUAAAUAUUUGUGGAGCAC-------------GGUGUGGACAUGAAAAUGGUAUCAUAGCACGCUUUUCACACAGCCGUGUGAAAAG-UACACACC- -...........((((((.(((........)))))).))).-------------((((((..(((....)))............(((((((((((....))))))))))-).))))))- ( -32.70, z-score = -2.02, R) >droAna3.scaffold_12916 11291584 105 + 16180835 -UGGCAUGCCUUUGUAAAAUGGUUAAAUAUUUGAGUGUAGGGAAAA-----------AUGUUAAUUAAAAUGGUAUC-UAGCACGCUUUUCACAUGGACCUGUGAAAAAAUAAACAUU- -((((((.((((....(((((......)))))......))))....-----------)))))).....((((((...-..))....((((((((......))))))))......))))- ( -15.00, z-score = 1.04, R) >dp4.chr4_group1 4837990 115 - 5278887 -UGGCAUUCCUUUGUCGAACAGUUAAAUAUUUGUAUAGAAUGGCUGUG--GCUGUGUGGAAAUGUGAAAAUGGUAUCAUAGCACGCUUUUCACAUGAGCGUGUGAAAAG-UACACAGAA -.((((......))))..(((((((..(((....)))...))))))).--.((((((......(((...(((....)))..)))(((((((((((....))))))))))-))))))).. ( -33.70, z-score = -2.26, R) >droPer1.super_1 9847081 117 - 10282868 -UGGCAUUCCUUUGUCGAACAGUUAAAUAUUUGUAUAGAAUGGCUGUGUGGAAAUGUGGAAAUGUGAAAAUGGUAUCAUAGCACGCUUUUCACAUGAGCGUGUGAAAAG-UACACAGAA -.((((......))))..(((((((..(((....)))...)))))))(((.....(((....(((((........))))).)))(((((((((((....))))))))))-).))).... ( -30.90, z-score = -1.75, R) >droWil1.scaffold_180708 4409206 106 - 12563649 UGACAUUCCUCA-AACAAACGGUUAAAUAUUUGUUAAGAAUGAGAA-----------AUGGAAAUGAAAAUUGUAUCAUAGCACGCUUUUCACAUGAGCGUGUGAAAGG-CACACAUAG ...((((.((((-....(((((........))))).....)))).)-----------)))(..((((........))))..)..(((((((((((....))))))))))-)........ ( -24.60, z-score = -2.26, R) >droVir3.scaffold_12963 16044460 90 - 20206255 UGGCAUUCCUAAAGGUAAACAGUUAAAUAUUUGUUUAGAAUUGGA-------------------------UCGUAUCGUAGCACGCUUUUCACAUGAGCGUGUGAAAAG-CAUACA--- ..((..(((......(((((((........))))))).....)))-------------------------..))...(((....(((((((((((....))))))))))-).))).--- ( -24.50, z-score = -2.73, R) >droGri2.scaffold_15252 1245954 90 + 17193109 ---GCAUUCUGAUGUCAAACAGUUAAAUAUUUGUGUAGACU-------------------------GGGAUCAUAUCCCAGCACGCUUUUCACAUGAGCGUGUGAAAAG-CACACAAUA ---.....(((........)))........((((((.(.((-------------------------(((((...))))))))..(((((((((((....))))))))))-))))))).. ( -31.70, z-score = -4.55, R) >droMoj3.scaffold_6500 24325083 84 - 32352404 --------UUAAUGGAAAACAGUUAAAUAUUUGUU-AGAACUGGA-------------------------UCGUAUCAUAGCACGCUUUUCACAUGAGUGUGUGAAAAG-CACACACAA --------......((...(((((...((.....)-).)))))..-------------------------))............(((((((((((....))))))))))-)........ ( -19.90, z-score = -1.99, R) >consensus _UGGCAUUCCUCUGUCAAACGGUUAAAUAUUUGUGUAGAAC_____________UGUGUGGACAUGAAAAUGGUAUCAUAGCACGCUUUUCACAUGAGCGUGUGAAAAG_UACACACCA ..................((((........))))...................................................((((((((((....)))))))))).......... (-10.97 = -10.62 + -0.35)

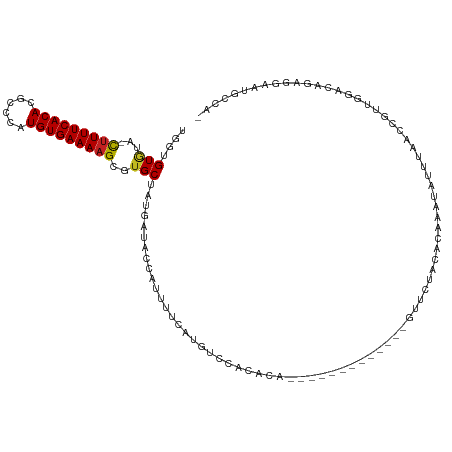
| Location | 5,881,057 – 5,881,161 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 69.29 |
| Shannon entropy | 0.64233 |
| G+C content | 0.40537 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -9.03 |
| Energy contribution | -9.05 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
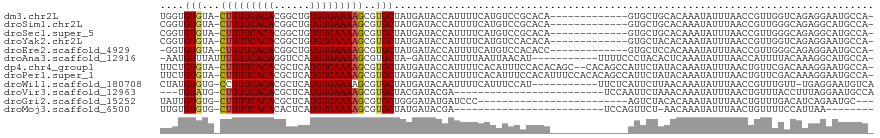
>dm3.chr2L 5881057 104 - 23011544 UGGUGUGUA-CUUUUCACACGGCUGUGUGAAAAGCGUGCUAUGAUACCAUUUUCAUGUCCGCACA-------------GUGCUGCACAAAUAUUUAACCGUUGGUCAGAGGAAUGCCA- ..(((((((-((((((((((....)))))))))(((..(.((((........)))))..)))...-------------)))).))))..............(((.((......)))))- ( -29.70, z-score = -0.55, R) >droSim1.chr2L 5671616 104 - 22036055 CGGUGUGUA-CUUUUCACACGGCUGUGUGAAAAGCGUGCUAUGAUACCAUUUUCAUGUCCGCACA-------------GUGCUGCACAAAUAUUUAACCGUUGGGCAGAGGCAUGCCA- .(((((((.-((((((((((....)))))))))).(((((((((........)))))...)))).-------------...((((.(((...........))).))))..))))))).- ( -34.60, z-score = -1.32, R) >droSec1.super_5 3950893 104 - 5866729 CGGUGUGUA-CUUUUCACACGGCUGUGUGAAAAGCGUGCUAUGAUACCAUUUUCAUGUCCGCACA-------------GUGCUGCACAAAUAUUUAACCGUUGGGCAGAGGCAUGCCA- .(((((((.-((((((((((....)))))))))).(((((((((........)))))...)))).-------------...((((.(((...........))).))))..))))))).- ( -34.60, z-score = -1.32, R) >droYak2.chr2L 9007980 104 + 22324452 CGGUGUGUA-CUUUUCACACGGCUGUGUGAAAAGCGUGCUAUGAUACCAUUUUCAUGUCCACACA-------------GUGCUACACAAAUAUUUAACCGUUGGUCAGAGGAAUGCCA- .((((((((-((((((((((....)))))))))).(((.(((((........)))))..)))...-------------....))))).....(((.(((...))).))).....))).- ( -27.10, z-score = -0.30, R) >droEre2.scaffold_4929 5964883 103 - 26641161 -GGUGUGUA-CUUUUCACACGGCUGUGUGAAAAGCGUGCUAUGAUACCAUUUUCAUGUCCACACC-------------GUGCUCCACAAAUAUUUAACCGUUGGGCAGAGGAAUGCCA- -((((((..-((((((((((....))))))))))((((..(((....)))...))))..))))))-------------(((...)))................((((......)))).- ( -31.40, z-score = -1.47, R) >droAna3.scaffold_12916 11291584 105 - 16180835 -AAUGUUUAUUUUUUCACAGGUCCAUGUGAAAAGCGUGCUA-GAUACCAUUUUAAUUAACAU-----------UUUUCCCUACACUCAAAUAUUUAACCAUUUUACAAAGGCAUGCCA- -..........((((((((......))))))))(((((((.-....................-----------....................................)))))))..- ( -14.62, z-score = -0.79, R) >dp4.chr4_group1 4837990 115 + 5278887 UUCUGUGUA-CUUUUCACACGCUCAUGUGAAAAGCGUGCUAUGAUACCAUUUUCACAUUUCCACACAGC--CACAGCCAUUCUAUACAAAUAUUUAACUGUUCGACAAAGGAAUGCCA- ..((((((.-(((((((((......))))))))).(((..(((....)))...)))......)))))).--....(.((((((...(.((((......)))).).....)))))).).- ( -22.60, z-score = -1.69, R) >droPer1.super_1 9847081 117 + 10282868 UUCUGUGUA-CUUUUCACACGCUCAUGUGAAAAGCGUGCUAUGAUACCAUUUUCACAUUUCCACAUUUCCACACAGCCAUUCUAUACAAAUAUUUAACUGUUCGACAAAGGAAUGCCA- ..((((((.-(((((((((......))))))))).(((..(((....)))...)))..............)))))).((((((...(.((((......)))).).....))))))...- ( -21.80, z-score = -1.91, R) >droWil1.scaffold_180708 4409206 106 + 12563649 CUAUGUGUG-CCUUUCACACGCUCAUGUGAAAAGCGUGCUAUGAUACAAUUUUCAUUUCCAU-----------UUCUCAUUCUUAACAAAUAUUUAACCGUUUGUU-UGAGGAAUGUCA ....(..((-(.(((((((......))))))).)))..).((((........))))...(((-----------((((((.....(((((((........)))))))-)))))))))... ( -27.10, z-score = -3.68, R) >droVir3.scaffold_12963 16044460 90 + 20206255 ---UGUAUG-CUUUUCACACGCUCAUGUGAAAAGCGUGCUACGAUACGA-------------------------UCCAAUUCUAAACAAAUAUUUAACUGUUUACCUUUAGGAAUGCCA ---.(((((-(((((((((......))))))))))))))..........-------------------------(((((...((((((..........))))))...)).)))...... ( -23.40, z-score = -3.94, R) >droGri2.scaffold_15252 1245954 90 - 17193109 UAUUGUGUG-CUUUUCACACGCUCAUGUGAAAAGCGUGCUGGGAUAUGAUCCC-------------------------AGUCUACACAAAUAUUUAACUGUUUGACAUCAGAAUGC--- ..(((((((-(((((((((......))))))))))..((((((((...)))))-------------------------)))..))))))........(((........))).....--- ( -31.40, z-score = -4.38, R) >droMoj3.scaffold_6500 24325083 84 + 32352404 UUGUGUGUG-CUUUUCACACACUCAUGUGAAAAGCGUGCUAUGAUACGA-------------------------UCCAGUUCU-AACAAAUAUUUAACUGUUUUCCAUUAA-------- ....(..((-(((((((((......)))))))))))..)..((((..((-------------------------..(((((..-...........)))))..))..)))).-------- ( -21.02, z-score = -3.10, R) >consensus UGGUGUGUA_CUUUUCACACGCCCAUGUGAAAAGCGUGCUAUGAUACCAUUUUCAUGUCCACACA_____________GUUCUACACAAAUAUUUAACCGUUGGACAGAGGAAUGCCA_ ....(((...(((((((((......)))))))))..)))................................................................................ ( -9.03 = -9.05 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:20:03 2011