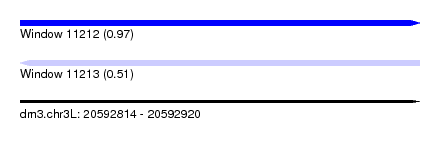
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,592,814 – 20,592,920 |
| Length | 106 |
| Max. P | 0.965614 |

| Location | 20,592,814 – 20,592,920 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 87.39 |
| Shannon entropy | 0.27624 |
| G+C content | 0.64884 |
| Mean single sequence MFE | -47.01 |
| Consensus MFE | -34.58 |
| Energy contribution | -35.98 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20592814 106 + 24543557 ---CUUCCACCACCCGCCUGCUGGCUAUUGUGGUGCGCCGCCAUCGCAGCCACCGCCUGCUGCUGCUGCUCCUGCAGCAGACAGUGGAUCUUGAACCAGUUGGAGCGAC ---...........((((((((((.....((((((.((.((....)).))))))))(..((((((((((....))))))).)))..)........)))).))).))).. ( -45.70, z-score = -2.60, R) >droSec1.super_11 482639 106 + 2888827 ---CUUCCGCCACCCGCCUGCUGGCUAUUGUGGUGCGCCGCCAUCGCAGCCACCGCCUGCUGCUGCUGCUCCUGCAGCAGACAGUGGAUCUUGAACCAGUUGGAGCGAC ---....(((......((.(((((.....((((((.((.((....)).))))))))(..((((((((((....))))))).)))..)........))))).)).))).. ( -46.80, z-score = -2.56, R) >droYak2.chr3L 4238938 106 - 24197627 ---CUGCCGCCACCCGCCUGCUGGCUAUUGUGGUGCGCCGCCAUCGCAGCCACCGCCUGUUGCUGCUGCUCCUGCAGCAGACAGUGGAUCUUGAACCAGUUGGAGCGAC ---....(((......((.(((((.....((((((.((.((....)).))))))))(..((((((((((....))))))).)))..)........))))).)).))).. ( -44.00, z-score = -1.44, R) >droEre2.scaffold_4784 20287022 106 + 25762168 ---CCUCCGCCUCCCGCCUGCUGGCUAUUGUGGUGCGCCGCCAUCGCAGCCACCGCCUGCUGCUGCUGCUCCUGCAGCAGACAGUGGAUCUUGAACCAGUUGGAGCGAC ---....(((.(((.....(((((.....((((((.((.((....)).))))))))(..((((((((((....))))))).)))..)........))))).)))))).. ( -48.20, z-score = -3.05, R) >droAna3.scaffold_13337 10944760 109 + 23293914 ACUCCACCUCCUCCACCCUGCUGGCUGUUGUGGUGCGCCGCCAUCGCUGCCACCGCCUGCUGCUGCUGCUCCUGGAGGAGGCAGUGGAUCUUGAACCAGUUGGAGCGCC .((((((((((((((....((.(((.((.((((((.((.((....)).))))))))..)).)))...))...)))))))))...(((........)))..))))).... ( -45.90, z-score = -1.51, R) >droWil1.scaffold_180949 2273242 97 + 6375548 ---CCGCCGCCAGCCGCCUGCUGA---------UGAGCCUGCAUGGCGGCCACCGCCUGUUGCUGCUGCUCCUGCAGCAGACAGUGAAUCUUGAACCAAUUGGAGCGAC ---..(((((((...((..(((..---------..)))..)).)))))))...(((((((..(((((((....)))))))))))...........((....)).))).. ( -39.60, z-score = -2.34, R) >droSim1.chr3L 19944290 106 + 22553184 ---CUUCCGCCACCCGCCUGCUGGCUAUUGUGGUGCGCCGCCAUCGCAGCCACCGCCUGCUGCUGCUGCUCUUGCAGCAGACAGUGGAUCUUGAACCAGUUAGAGCGAC ---...........((((((((((.....((((((.((.((....)).))))))))(..((((((((((....))))))).)))..)........)))).))).))).. ( -47.00, z-score = -3.09, R) >droVir3.scaffold_13049 6478827 106 - 25233164 ---CCUCCGGCGCCAGCCUGCUGACUGUUGUGGUGUGCUGCCAUCGCCGCGACCGCCUGCUGCUGCUGCUCCUGCAGCAGACAGUGUAUCUUGAACCAGUUGGAGCGUC ---.((((((((.(((....)))...(((((((((((....)).))))))))))))).(((((((((((....))))))).))))................)))).... ( -45.40, z-score = -2.00, R) >droMoj3.scaffold_6680 12473783 106 + 24764193 ---CCCGCCGCGCCCGCCUGCUGGCUGUUGUGGUGUGCCGCCAUGGCAGCCACCGCCUGCUGCUGCUGCUCCUGCAGCAGGCAGUGGAUCUUGAACCAAUUGGAGCGGC ---...(((((..((((..(.(((((((..(((((...)))))..))))))).)(((((((((..........))))))))).))))........((....)).))))) ( -57.80, z-score = -3.09, R) >droGri2.scaffold_15110 5337870 106 - 24565398 ---GUCGCUCCGCCAGCCUGCUGGCUGUUGUGGUGUGCGGCCAUCGCAGCAACCGCCUGUUGCUGUUGCUCCUGCAGCAGACAGUGGAUCUUGAACCAGUUGGAGCGUC ---..(((((((((((....)))))((((((((((......)))))))))).((((.(((..(((((((....))))))))))))))..............)))))).. ( -49.70, z-score = -2.48, R) >consensus ___CCUCCGCCACCCGCCUGCUGGCUAUUGUGGUGCGCCGCCAUCGCAGCCACCGCCUGCUGCUGCUGCUCCUGCAGCAGACAGUGGAUCUUGAACCAGUUGGAGCGAC ................((.(((((.....((((((.((.((....)).))))))))(((((((((((((....))))))).))))))........))))).))...... (-34.58 = -35.98 + 1.40)

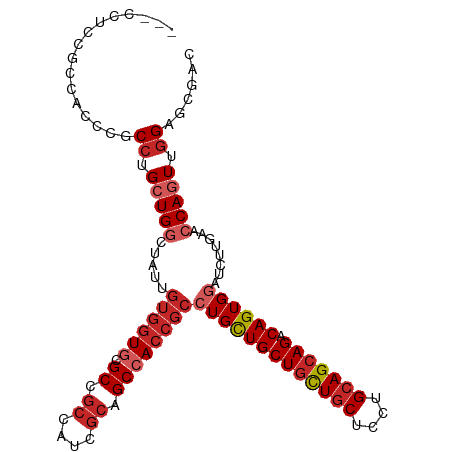
| Location | 20,592,814 – 20,592,920 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.39 |
| Shannon entropy | 0.27624 |
| G+C content | 0.64884 |
| Mean single sequence MFE | -48.15 |
| Consensus MFE | -33.23 |
| Energy contribution | -33.70 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20592814 106 - 24543557 GUCGCUCCAACUGGUUCAAGAUCCACUGUCUGCUGCAGGAGCAGCAGCAGCAGGCGGUGGCUGCGAUGGCGGCGCACCACAAUAGCCAGCAGGCGGGUGGUGGAAG--- ((((((((....)).....(..((((((((((((((..........))))))))))))))...)...)))))).((((((....(((....)))..))))))....--- ( -50.20, z-score = -2.00, R) >droSec1.super_11 482639 106 - 2888827 GUCGCUCCAACUGGUUCAAGAUCCACUGUCUGCUGCAGGAGCAGCAGCAGCAGGCGGUGGCUGCGAUGGCGGCGCACCACAAUAGCCAGCAGGCGGGUGGCGGAAG--- (((((.((...((((.(......(((((((((((((..........)))))))))))))(((((....)))))).)))).....(((....)))))))))).....--- ( -49.90, z-score = -1.79, R) >droYak2.chr3L 4238938 106 + 24197627 GUCGCUCCAACUGGUUCAAGAUCCACUGUCUGCUGCAGGAGCAGCAGCAACAGGCGGUGGCUGCGAUGGCGGCGCACCACAAUAGCCAGCAGGCGGGUGGCGGCAG--- ((((((((..((((((.........(((((((((((....)))))))..))))(.(((((((((....))))).)))).)...))))))...).))))))).....--- ( -48.80, z-score = -1.51, R) >droEre2.scaffold_4784 20287022 106 - 25762168 GUCGCUCCAACUGGUUCAAGAUCCACUGUCUGCUGCAGGAGCAGCAGCAGCAGGCGGUGGCUGCGAUGGCGGCGCACCACAAUAGCCAGCAGGCGGGAGGCGGAGG--- .(((((((...((((.(......(((((((((((((..........)))))))))))))(((((....)))))).)))).....(((....))).))).))))...--- ( -49.00, z-score = -1.58, R) >droAna3.scaffold_13337 10944760 109 - 23293914 GGCGCUCCAACUGGUUCAAGAUCCACUGCCUCCUCCAGGAGCAGCAGCAGCAGGCGGUGGCAGCGAUGGCGGCGCACCACAACAGCCAGCAGGGUGGAGGAGGUGGAGU ...(((((...(((........)))..((((((((((...((....)).((.(((.((((..(((.......))).))))....))).))....))))))))))))))) ( -46.10, z-score = -0.59, R) >droWil1.scaffold_180949 2273242 97 - 6375548 GUCGCUCCAAUUGGUUCAAGAUUCACUGUCUGCUGCAGGAGCAGCAGCAACAGGCGGUGGCCGCCAUGCAGGCUCA---------UCAGCAGGCGGCUGGCGGCGG--- (((((.((....)).........((((((((((((...((((....(((...(((((...))))).)))..)))).---------.)))))))))).)))))))..--- ( -43.10, z-score = -1.42, R) >droSim1.chr3L 19944290 106 - 22553184 GUCGCUCUAACUGGUUCAAGAUCCACUGUCUGCUGCAAGAGCAGCAGCAGCAGGCGGUGGCUGCGAUGGCGGCGCACCACAAUAGCCAGCAGGCGGGUGGCGGAAG--- (((((((....((((.(......(((((((((((((..........)))))))))))))(((((....)))))).)))).....(((....)))))))))).....--- ( -50.20, z-score = -2.29, R) >droVir3.scaffold_13049 6478827 106 + 25233164 GACGCUCCAACUGGUUCAAGAUACACUGUCUGCUGCAGGAGCAGCAGCAGCAGGCGGUCGCGGCGAUGGCAGCACACCACAACAGUCAGCAGGCUGGCGCCGGAGG--- ....((((.................(((.(((((((....))))))))))..((((((.((.((....)).))..)))......(((((....)))))))))))).--- ( -42.30, z-score = -0.78, R) >droMoj3.scaffold_6680 12473783 106 - 24764193 GCCGCUCCAAUUGGUUCAAGAUCCACUGCCUGCUGCAGGAGCAGCAGCAGCAGGCGGUGGCUGCCAUGGCGGCACACCACAACAGCCAGCAGGCGGGCGCGGCGGG--- ((((((((...((((........(((((((((((((..........)))))))))))))(((((....)))))..)))).....(((....)))))).)))))...--- ( -57.90, z-score = -3.01, R) >droGri2.scaffold_15110 5337870 106 + 24565398 GACGCUCCAACUGGUUCAAGAUCCACUGUCUGCUGCAGGAGCAACAGCAACAGGCGGUUGCUGCGAUGGCCGCACACCACAACAGCCAGCAGGCUGGCGGAGCGAC--- ..((((((...((((.........((((((((.(((.(......).))).))))))))(((.((....)).))).)))).....(((((....)))))))))))..--- ( -44.00, z-score = -1.67, R) >consensus GUCGCUCCAACUGGUUCAAGAUCCACUGUCUGCUGCAGGAGCAGCAGCAGCAGGCGGUGGCUGCGAUGGCGGCGCACCACAAUAGCCAGCAGGCGGGUGGCGGAAG___ ..((((((..((((((.........(((((((((((....)))))))..))))..((((((.((....)).)).)))).....))))))..))..)))).......... (-33.23 = -33.70 + 0.47)
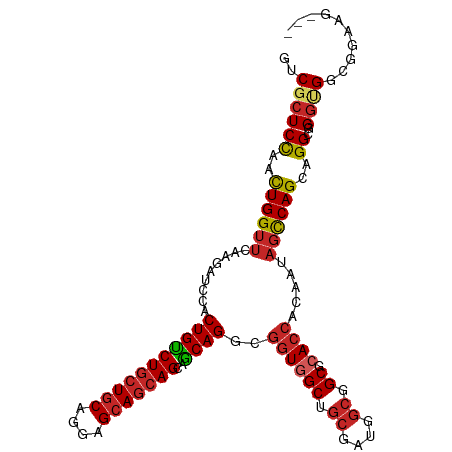
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:21 2011