| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,583,461 – 20,583,561 |
| Length | 100 |
| Max. P | 0.933384 |

| Location | 20,583,461 – 20,583,556 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 51.98 |
| Shannon entropy | 0.94251 |
| G+C content | 0.47439 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -8.00 |
| Energy contribution | -8.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
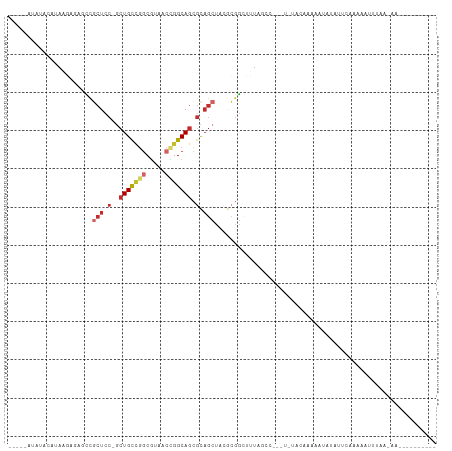
>dm3.chr3L 20583461 95 + 24543557 -----AUAUACAUAAGAACGCCGCUCC-GCUGCUGGCGUACCCGGCAGCGCAGCUACGCGG-AUUAGCC---UAAGUCCAAAUAUAUAAAAAACUGUAAAAUCAG------- -----...((((.......((.(((.(-((((((((.....))))))))).)))...))((-(((....---..)))))...............)))).......------- ( -29.00, z-score = -2.70, R) >droSim1.chr3h_random 884955 77 + 1452968 -----AAACGCAAAUAAGCGUCGCUGC-GCUGCAGGCUAUGCCGGCAGCCCUGCUCCUCGACUUAGGC--------UAAGAAACCAUUGUA--------------------- -----....((...((((((..((.(.-(((((.(((...))).))))).).))....)).)))).))--------...............--------------------- ( -21.80, z-score = 0.28, R) >droYak2.chrX_random 1263061 86 - 1802292 ------------UAAGAGAGCCGCUCC-GCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGG-CUUAGCC---UUCGACGAAAUAUAAUUAAGAAUCACAAAAC--------- ------------.....(((((((...-((((((((((....)))))..)))))...))))-)))....---...............................--------- ( -26.00, z-score = -1.50, R) >droEre2.scaffold_4855 234983 88 - 242605 -----------AUAAGAGAGCCGCUCC-GCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGU---UUAGCCGAAAUAUACUUAAGAAUGUAUCGCC--------- -----------.((((((((((((..(-(((((..((...))..)))))).......)))))))).((.---....))........)))).............--------- ( -28.80, z-score = -0.23, R) >droAna3.scaffold_13767 1524744 91 + 1685616 -----ACAUAAGUAAUAAAGUCUCUCC-GCUGCCGGCUUCAAGGGCAGCGCAGCUACGAGGCUCUAAUAAAAUAUGGAAUAAGAUAUUCAUUGUUAA--------------- -----.((((..((.((.((((((...-((((((.(((.....))).).)))))...)))))).)).))...))))(((((...)))))........--------------- ( -23.30, z-score = -1.51, R) >droPer1.super_1295 3566 103 + 6930 GAUAAGAACACAUAGUUCUGCCGCUCC-GCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGGCUUUGGGC---U-UACAAAAGCCCAAACCACAAUUGUAAAAUAUAGA---- ....(((((.....)))))((.(((.(-((((((((.....))))))))).)))...))((.(((((((---(-(....)))))))))))..................---- ( -42.00, z-score = -4.49, R) >droWil1.scaffold_181143 551653 103 + 1323777 -----UUAUUCAUAAGCAGACUGCUCC-UCUGUCGGCGUCUCUGGCAGCGUAACUACACGGUCUUUAGC---UCUACAAGACAAUUGUUAAAGAAUUAAGUAUAUUGAAAAA -----...((((((.((((((((....-.(((((((.....))))))).((....)).))))))((((.---(((((((.....))))...))).)))))).)).))))... ( -17.90, z-score = 0.58, R) >droVir3.scaffold_12941 339862 80 - 746104 -----AUAUACAAAAGACUCUCUCUCCCGCUGCUUGCGUAACCGGCACAGCAGCUAUGCGGUCUUAGC------CAUAAGAAUAUAUUGAA--------------------- -----.....(((...............(((((((((.......))).))))))((((...(((((..------..))))).)))))))..--------------------- ( -16.30, z-score = -0.25, R) >droMoj3.scaffold_6496 24532205 94 + 26866924 -----AUAUACAUAAGCGGACCGCUGC-UCUGCCGGCGUCACUGGCAGCGCAGCCA-ACCGAUUUUAAC---UGUACUUGACAAUUUUCAUAGAUUUAAGAAUU-------- -----...((((.((((((...(((((-.(((((((.....))))))).)))))..-.))).)))....---))))(((((..((((....)))))))))....-------- ( -28.00, z-score = -2.98, R) >consensus _____AUAUACAUAAGAGAGCCGCUCC_GCUGCCGGCGUAACCGGCAGCGCAGCUACGCGGCUUUAGCC___U_UACAAAAAUAUAUUCAAAAAUUUAA_AA__________ ......................(((....(((((((.....)))))))...))).......................................................... ( -8.00 = -8.67 + 0.67)


| Location | 20,583,461 – 20,583,561 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 51.74 |
| Shannon entropy | 1.02542 |
| G+C content | 0.47145 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -8.00 |
| Energy contribution | -8.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
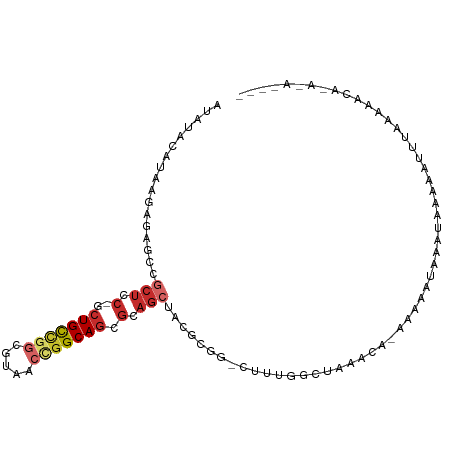
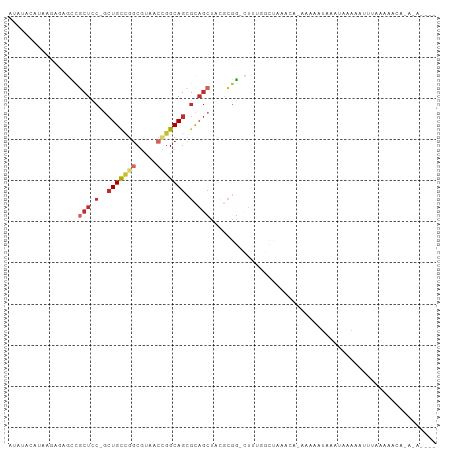
>dm3.chr3L 20583461 100 + 24543557 AUAUACAUAAGAACGCCGCUCC-GCUGCUGGCGUACCCGGCAGCGCAGCUACGCGG-AUUAGCCUAAGUCCAAAUAUAUAAAAAACUGUAAAAUCAGAGAGA-- (((((.........((.(((.(-((((((((.....))))))))).)))...))((-(((......)))))...)))))......(((......))).....-- ( -29.80, z-score = -2.36, R) >droSim1.chr3h_random 884955 77 + 1452968 AAACGCAAAUAAGCGUCGCUGC-GCUGCAGGCUAUGCCGGCAGCCCUGCUCCUCGA-CUUAGGCUAAG--------AAACCAUUGUA----------------- ....((...((((((..((.(.-(((((.(((...))).))))).).))....)).-)))).))....--------...........----------------- ( -21.80, z-score = 0.28, R) >droYak2.chrX_random 1263061 91 - 1802292 -------UAAGAGAGCCGCUCC-GCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGG-CUUAGCCUUCGACGAAAUAUAAUUAAGAAUCACAAAACAGACA---- -------.....(((((((...-((((((((((....)))))..)))))...))))-)))........................................---- ( -26.00, z-score = -1.24, R) >droEre2.scaffold_4855 234983 93 - 242605 ------AUAAGAGAGCCGCUCC-GCUGCUGGCGUAGCUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCCGAAAUAUACUUAAGAAUGUAUCGCCAGAUA---- ------.((((((((((((..(-(((((..((...))..)))))).......)))))))).((.....))........))))......((((....))))---- ( -29.90, z-score = -0.28, R) >droAna3.scaffold_13767 1524744 96 + 1685616 ACAUAAGUAAUAAAGUCUCUCC-GCUGCCGGCUUCAAGGGCAGCGCAGCUACGAGG-CUCUAAUAAAAUAUGGAAUAAGAUAUUCAUUGUUAAAAACG------ .((((..((.((.((((((...-((((((.(((.....))).).)))))...))))-)).)).))...))))(((((...))))).............------ ( -23.30, z-score = -1.66, R) >droPer1.super_1295 3571 101 + 6930 GAACACAUAGUUCUGCCGCUCC-GCUGCCGGCGUCACUGGCAGCGCAGCUCCGCGG-CUUUGGGCUUACAAAAGCCCAAACCACAAUUGUAAAAUAUAGAAGA- ((((.....)))).((.(((.(-((((((((.....))))))))).)))...))((-.(((((((((....))))))))))).....................- ( -40.60, z-score = -3.97, R) >droWil1.scaffold_181143 551653 103 + 1323777 UUAUUCAUAAGCAGACUGCUCC-UCUGUCGGCGUCUCUGGCAGCGUAACUACACGGUCUUUAGCUCUACAAGACAAUUGUUAAAGAAUUAAGUAUAUUGAAAAA ...((((((.((((((((....-.(((((((.....))))))).((....)).))))))((((.(((((((.....))))...))).)))))).)).))))... ( -17.90, z-score = 0.58, R) >droVir3.scaffold_12941 339862 80 - 746104 AUAUACAAAAGACUCUCUCUCCCGCUGCUUGCGUAACCGGCACAGCAGCUAUGCGG-UCUUAGCCAUA------AGAAUAUAUUGAA----------------- .....(((...............(((((((((.......))).))))))((((...-(((((....))------))).)))))))..----------------- ( -16.30, z-score = -0.25, R) >droMoj3.scaffold_6496 24532205 96 + 26866924 AUAUACAUAAGCGGACCGCUGC-UCUGCCGGCGUCACUGGCAGCGCAGCCA-ACCGAUUUUAACUGUACUUGACAAUUUUCAUAGAUUUAAGAAU--UAA---- ...((((.((((((...(((((-.(((((((.....))))))).)))))..-.))).)))....))))(((((..((((....)))))))))...--...---- ( -28.00, z-score = -2.92, R) >consensus AUAUACAUAAGAGAGCCGCUCC_GCUGCCGGCGUAACCGGCAGCGCAGCUACGCGG_CUUUGGCUAAACA_AAAAAUAAAUAAAAAUUUAAAAACA_A_A____ .................(((....(((((((.....)))))))...)))....................................................... ( -8.00 = -8.67 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:19 2011