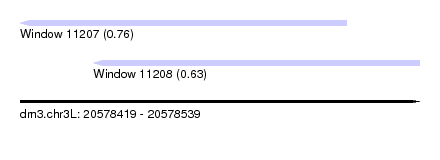
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,578,419 – 20,578,539 |
| Length | 120 |
| Max. P | 0.759579 |

| Location | 20,578,419 – 20,578,517 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.41 |
| Shannon entropy | 0.58288 |
| G+C content | 0.33571 |
| Mean single sequence MFE | -10.58 |
| Consensus MFE | -5.46 |
| Energy contribution | -4.85 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
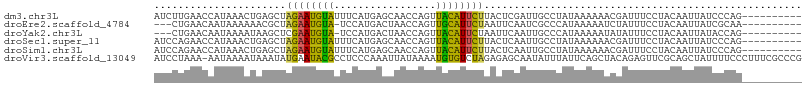
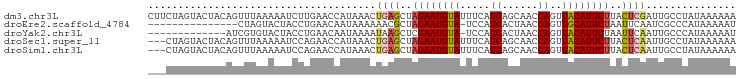
>dm3.chr3L 20578419 98 - 24543557 AUCUUGAACCAUAAACUGAGCUAGAAUGUAUUUCAUGAGCAACCAGUUACAUUCUUACUCGAUUGCCUAUAAAAAACGAUUUCCUACAAUUAUCCCAG---------- ................((((..((((((((.....((......))..))))))))..)))).....................................---------- ( -12.30, z-score = -1.21, R) >droEre2.scaffold_4784 20273096 94 - 25762168 ---CUGAACAAUAAAAAACGCUAGAAUGUA-UCCAUGACUAACCAGUUGCAUUCUAAUUCAAUCGCCCAUAAAAAUCUAUUUCCUACAAUUAUCGCAA---------- ---..................(((((((((-....((......))..)))))))))..........................................---------- ( -9.40, z-score = -1.23, R) >droYak2.chr3L 4225085 94 + 24197627 ---CUGAACAAUAAAAUAAGCUCGAAUGUA-UCCAUGACUAACCAGUUACAUUCUAAUUCAAUUGCCCAUAAAAAUAUAUUUCCUACAAUUAUACCAG---------- ---(((.................(((((((-....((......))..)))))))......(((((.....................)))))....)))---------- ( -7.40, z-score = -0.83, R) >droSec1.super_11 468704 98 - 2888827 AUCCAGAACCAUAAACUGAGCUAGAAUGUAUUUCAUGAGCAACCAGUUACAUUCUUACUCAAUUGCCUAUAAAAAACGAUUUCCUACAAUUAUCCCAG---------- ................((((..((((((((.....((......))..))))))))..)))).....................................---------- ( -13.00, z-score = -2.01, R) >droSim1.chr3L 19930390 98 - 22553184 AUCCAGAACCAUAAACUGAGCUAGAAUGUAUUUCAUGAGCAACCAGUUACAUUCUUACUCAAUUGCCUAUAAAAAACGAUUUCCUACAAUUAUCCCAG---------- ................((((..((((((((.....((......))..))))))))..)))).....................................---------- ( -13.00, z-score = -2.01, R) >droVir3.scaffold_13049 6465612 107 + 25233164 AUCCUAAA-AAUAAAAUAAAUAUGAAUACGCCUCCCAAAUUAUAAAAUGUGUCUAGAGAGCAAUAUUUAUUCAGCUACAGAGUUCGCAGCUAUUUUCCCUUUCGCCCG ........-.....(((((((((......(((((....((........)).....))).)).))))))))).((((...(....)..))))................. ( -8.40, z-score = 0.94, R) >consensus AUCCUGAACAAUAAAAUAAGCUAGAAUGUAUUUCAUGAGCAACCAGUUACAUUCUUACUCAAUUGCCUAUAAAAAACGAUUUCCUACAAUUAUCCCAG__________ ......................((((((((.....((......))..))))))))..................................................... ( -5.46 = -4.85 + -0.61)
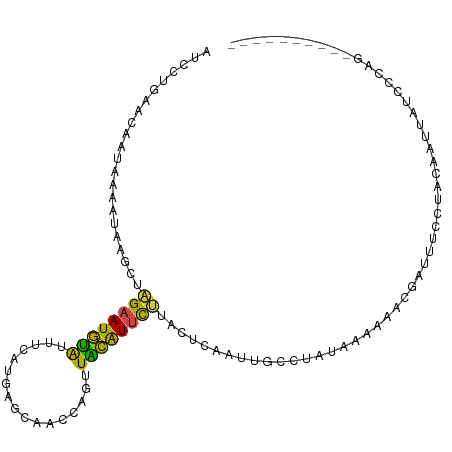
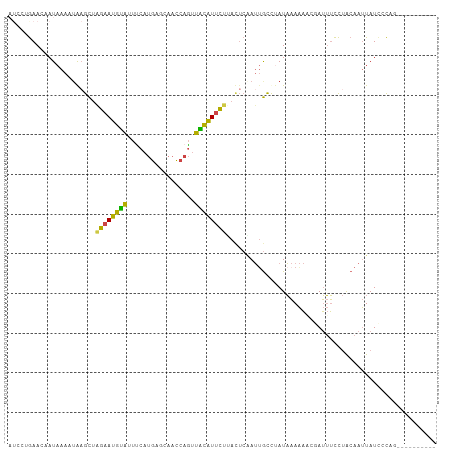
| Location | 20,578,441 – 20,578,539 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 76.13 |
| Shannon entropy | 0.38717 |
| G+C content | 0.32216 |
| Mean single sequence MFE | -11.70 |
| Consensus MFE | -8.62 |
| Energy contribution | -9.10 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20578441 98 - 24543557 CUUCUAGUACUACAGUUUAAAAAUCUUGAACCAUAAACUGAGCUAGAAUGUAUUUCAUGAGCAACCAGUUACAUUCUUACUCGAUUGCCUAUAAAAAA ......(((.....((((((.....)))))).......((((..((((((((.....((......))..))))))))..))))..))).......... ( -14.60, z-score = -0.97, R) >droEre2.scaffold_4784 20273118 82 - 25762168 ---------------CUAGUACUACCUGAACAAUAAAAAACGCUAGAAUGUA-UCCAUGACUAACCAGUUGCAUUCUAAUUCAAUCGCCCAUAAAAAU ---------------............................(((((((((-....((......))..))))))))).................... ( -9.40, z-score = -1.75, R) >droYak2.chr3L 4225107 84 + 24197627 -------------AUCGUGUACUACCUGAACAAUAAAAUAAGCUCGAAUGUA-UCCAUGACUAACCAGUUACAUUCUAAUUCAAUUGCCCAUAAAAAU -------------....(((.(.....).))).............(((((((-....((......))..)))))))...................... ( -7.20, z-score = -0.39, R) >droSec1.super_11 468726 95 - 2888827 ---CUAGUACUACAGUUUAAAAAUCCAGAACCAUAAACUGAGCUAGAAUGUAUUUCAUGAGCAACCAGUUACAUUCUUACUCAAUUGCCUAUAAAAAA ---...(((....((((((..............))))))(((..((((((((.....((......))..))))))))..)))...))).......... ( -13.64, z-score = -1.20, R) >droSim1.chr3L 19930412 95 - 22553184 ---CUAGUACUACAGUUUAAAAAUCCAGAACCAUAAACUGAGCUAGAAUGUAUUUCAUGAGCAACCAGUUACAUUCUUACUCAAUUGCCUAUAAAAAA ---...(((....((((((..............))))))(((..((((((((.....((......))..))))))))..)))...))).......... ( -13.64, z-score = -1.20, R) >consensus ___CUAGUACUACAGUUUAAAAAUCCUGAACCAUAAACUGAGCUAGAAUGUAUUUCAUGAGCAACCAGUUACAUUCUUACUCAAUUGCCUAUAAAAAA ......................................((((..((((((((.....((......))..))))))))..))))............... ( -8.62 = -9.10 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:17 2011