
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,566,512 – 20,566,584 |
| Length | 72 |
| Max. P | 0.977030 |

| Location | 20,566,512 – 20,566,584 |
|---|---|
| Length | 72 |
| Sequences | 7 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 84.54 |
| Shannon entropy | 0.26975 |
| G+C content | 0.40355 |
| Mean single sequence MFE | -12.09 |
| Consensus MFE | -11.25 |
| Energy contribution | -11.19 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.977030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20566512 72 - 24543557 ACCACAUGACACAC----------AGCAAUAAUUACCCACUUGCCUGCACUUGAUAUUAAAAUCAGACAAAAAGGUAAGGCC------ ..............----------...............(((((((....(((((......)))))......)))))))...------ ( -11.20, z-score = -1.33, R) >droSim1.chrU 15032342 72 - 15797150 ACCACAUGACACAC----------AACAAUAAUUACCCACUUGCCUGCACUUGAUAUUAAAAUCAGACAAAAAGGUAAGGCC------ ..............----------...............(((((((....(((((......)))))......)))))))...------ ( -11.20, z-score = -1.83, R) >droSec1.super_11 456933 72 - 2888827 ACCACAUGACACAC----------AACAAUAAUUACCCACUUGCCUGCACUUGAUAUUAAAAUCAGACAAAAAGGUAAGGCC------ ..............----------...............(((((((....(((((......)))))......)))))))...------ ( -11.20, z-score = -1.83, R) >droYak2.chr3L 4211483 68 + 24197627 ACCACAUGACACAC-------------AAUAAUUACCCACUUGCCUGCACUUGAUAUUAAAAUCAGAC-AAAAGGUACGGCC------ ..............-------------............(.(((((....(((((......)))))..-...))))).)...------ ( -7.70, z-score = -0.34, R) >droEre2.scaffold_4784 20260914 69 - 25762168 ACCACAUGACACAC-------------AAUAAUUACCCACUUGCCUGCACUUGAUAUUAAAAUCAGACAGAAAGGCAAGACC------ ..............-------------............(((((((....(((((......)))))......)))))))...------ ( -13.00, z-score = -4.07, R) >droAna3.scaffold_13337 10919202 81 - 23293914 ACCACACGGCACACACAGUCAA-CAACAAUAAUUACCCACUUGCCUGCACUUGAUAUUAAAAUCAGACAAAAAGGUAAGGCC------ .......(((.......((...-..))............(((((((....(((((......)))))......))))))))))------ ( -14.00, z-score = -1.83, R) >droWil1.scaffold_180949 4130136 86 + 6375548 AUCAACUGGCACACACAGUCAACAAACAAUAAUUACCCACUUGCCUGCACUUGAUAUUAAAAUCAGAC--AAAGGCAAGCUCACUGCU ....((((.......))))....................(((((((....(((((......)))))..--..)))))))......... ( -16.30, z-score = -2.08, R) >consensus ACCACAUGACACAC__________AACAAUAAUUACCCACUUGCCUGCACUUGAUAUUAAAAUCAGACAAAAAGGUAAGGCC______ .......................................(((((((....(((((......)))))......)))))))......... (-11.25 = -11.19 + -0.06)

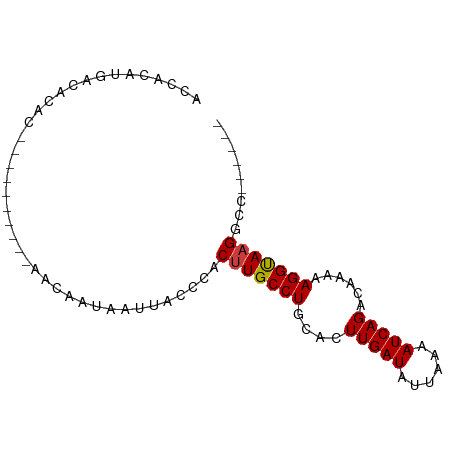
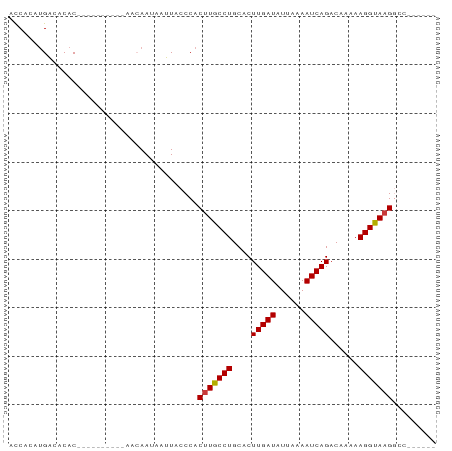
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:14 2011