
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,545,513 – 20,545,613 |
| Length | 100 |
| Max. P | 0.823030 |

| Location | 20,545,513 – 20,545,613 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 63.72 |
| Shannon entropy | 0.67714 |
| G+C content | 0.49776 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -10.85 |
| Energy contribution | -10.59 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20545513 100 + 24543557 ACACUUUAUAUAGGUAUAGGGUGCAGCAAAAGCAACAG------------CAACAGAAACAGGCGACAUCAACAUUG-CUGACGUAAACGAAGUCGUCUGCGCUCAAUCAUAG .(((((((((....))))))))).(((....((....)------------)........((((((((.((....(((-(....))))..)).)))))))).)))......... ( -25.00, z-score = -2.15, R) >droSec1.super_11 435945 100 + 2888827 ACACUUUAUAUAGGUAUAGGGUGCAGCAAGAGCAACAG------------CAACAGAAACAGGCGACAUCAACAUUG-CUGACGUAAACGAAGUCGUCUGCGCUCAAUCAUAG .(((((((((....)))))))))......((((..(((------------(((........(....).......)))-)))..(((.(((....))).)))))))........ ( -25.56, z-score = -2.01, R) >droYak2.chr3L 4188019 112 - 24197627 ACACUCUAUAUAGGUAUAGGGUGCAGCAAGAGCAACAGAAAGAGCAACAGCAACAGAAACAGGCGACAUCAACAUCG-CUGACGUAAACGAAGUCGUCUGCGCUCAAUCAUAG .(((((((((....)))))))))......((((..........((....))........((((((((.((.....((-....)).....)).)))))))).))))........ ( -31.50, z-score = -3.24, R) >droAna3.scaffold_13337 10898565 87 + 23293914 ACACUUUG-GUAGA---AGGGUGCAGCAGAAAAAGC--------------CAGCUC-------CAACAACGACAUCG-CUGACGUAAACGAAGUCGUCUGCGCUCAAUCAUAG ...((.((-((...---.((((((((.......((.--------------...)).-------.....(((((.(((-.(......).))).))))))))))))).)))).)) ( -19.60, z-score = 0.02, R) >droWil1.scaffold_180949 2219133 101 + 6375548 ---AUGGUAAAGGAAAUGAAGCGGGGUAGAGGGAGGGAGC---------GUGGCAUGGCAGGCGUUCUCUUCCAGCUUCCAACGUAAACGAAGUCAUCGUCGCUCAAUCAUAG ---............((((((((((((...((((((((((---------((.((...))..)))))))))))).(((((..........))))).))).)))))...)))).. ( -30.80, z-score = -1.41, R) >droVir3.scaffold_13049 6431284 83 - 25233164 ----UUCCACAGCAUAUAGGGUGCAGCCGGG-GAGCGA-------------------------CGAUGUGGACAACGCCUGACGUAAACGAAGUCGUCUGCGCUCAAUCAUAG ----..............(((((((((....-..))((-------------------------(((((((.....)))....((....))..)))))))))))))........ ( -21.60, z-score = 0.52, R) >droGri2.scaffold_15110 5292277 88 - 24565398 ACUCUUCGGUUACGUAUAGGGUGCAGCCGGGCGAGCGA-------------------------CGAUGUCGACAACGGCUGACGUAAACGAAGUCGUCUGCGCUCAAUCAUAG (((((.((....))...)))))......(((((...((-------------------------((((.(((...(((.....)))...))).))))))..)))))........ ( -28.10, z-score = -0.33, R) >consensus ACACUUUAUAUAGGUAUAGGGUGCAGCAGGAGCAGCAA____________CAACAG_____G_CGACAUCAACAUCG_CUGACGUAAACGAAGUCGUCUGCGCUCAAUCAUAG ..................(((((((((....................................(((........)))...(((.........)))).))))))))........ (-10.85 = -10.59 + -0.26)

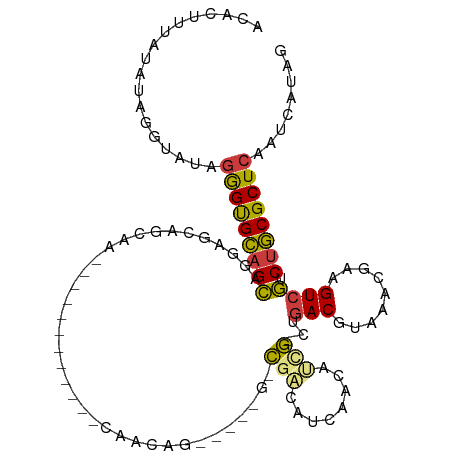
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:12 2011