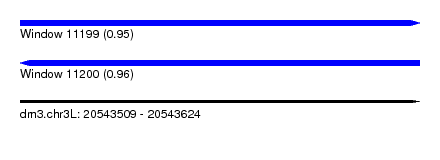
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,543,509 – 20,543,624 |
| Length | 115 |
| Max. P | 0.960776 |

| Location | 20,543,509 – 20,543,624 |
|---|---|
| Length | 115 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.03 |
| Shannon entropy | 0.34212 |
| G+C content | 0.36036 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -19.97 |
| Energy contribution | -19.82 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948655 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
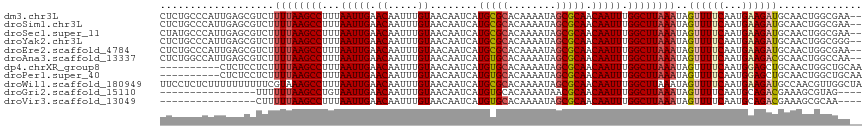
>dm3.chr3L 20543509 115 + 24543557 --UUCGCCAGUUGCAUCUUCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCGCAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAGACGCUCAAUGGGCAGAG --...(((((......)).((((((......((((((((.......(((((........)))))((((((..((.....)).))))))..))))))))......))))))))).... ( -31.30, z-score = -2.03, R) >droSim1.chr3L 19895263 115 + 22553184 --UUCGCCAGUUGCAUCUUCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCGCAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAGACGCUCAAUGGGCAGAG --...(((((......)).((((((......((((((((.......(((((........)))))((((((..((.....)).))))))..))))))))......))))))))).... ( -31.30, z-score = -2.03, R) >droSec1.super_11 433894 115 + 2888827 --UUCGCCAGUUGCAUCUUCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCGCAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAGACGCUCAAUGGGCAUAG --...(((((......)).((((((......((((((((.......(((((........)))))((((((..((.....)).))))))..))))))))......))))))))).... ( -31.30, z-score = -2.32, R) >droYak2.chr3L 4185931 115 - 24197627 --CCCGCCAGUUGCAUCUUCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCGCAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAGACGCUCAAUGGGCAGAG --...(((((......)).((((((......((((((((.......(((((........)))))((((((..((.....)).))))))..))))))))......))))))))).... ( -31.30, z-score = -2.02, R) >droEre2.scaffold_4784 20237476 115 + 25762168 --UUCGCCAGUUGCAUCUUCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCGCAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAGACGCUCAAUGGGCAGAG --...(((((......)).((((((......((((((((.......(((((........)))))((((((..((.....)).))))))..))))))))......))))))))).... ( -31.30, z-score = -2.03, R) >droAna3.scaffold_13337 10896738 115 + 23293914 --UUGGCCAGUUGCGUCUUCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCACAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAGACGCUCAAUGGCCAGAG --(((((((...(((((((.............(((((((....(((.((((......)))))))((((((..((.....)).))))))..))))))))))))))....))))))).. ( -40.11, z-score = -4.95, R) >dp4.chrXR_group8 7949421 107 + 9212921 UUGCAGCCAGUUGCAGCUCCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCACAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAGAGGAGAG---------- ((((((....))))))((((..(....)((.((((((((....(((.((((......)))))))((((((..((.....)).))))))..)))))))))).))))..---------- ( -29.70, z-score = -2.25, R) >droPer1.super_40 138919 107 + 810552 UUGCAGCCAGUUGCAGCUCCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCACAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAGAGGAGAG---------- ((((((....))))))((((..(....)((.((((((((....(((.((((......)))))))((((((..((.....)).))))))..)))))))))).))))..---------- ( -29.70, z-score = -2.25, R) >droWil1.scaffold_180949 4158078 117 - 6375548 UAGCCAACGUUGGCAUCUUCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCGCAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUUACGAAAAAAAAAAGAGAGGAA ..((((....)))).(((((.((...........(((((.......(((((........)))))((((((..((.....)).))))))..)))))............))..))))). ( -27.50, z-score = -1.59, R) >droGri2.scaffold_15110 19274813 97 + 24565398 ----CUACGCUUUCGUCUGCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGUUAUUUUGUGCACAUGAUUGUUACAAAUUGUUCAAUUACAGGCUUAAAAAA---------------- ----....((........))...........((((((((.(((((..(((....((((((.(((....))))))))).))).)))))...))))))))...---------------- ( -19.80, z-score = -1.21, R) >droVir3.scaffold_13049 18803745 97 + 25233164 ----UUGCGCUUUCGUCUGCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCACAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAAG---------------- ----....((........))........((.((((((((....(((.((((......)))))))((((((..((.....)).))))))..)))))))).))---------------- ( -23.00, z-score = -1.72, R) >consensus __UUCGCCAGUUGCAUCUUCAUUGAAAACUAUUUAAGCCAAAUUGUUGCGCUAUUUUGUGCGCAUGAUUGUUACAAAUUGUUCAAUUAAAGGCUUAAAAGACGCUCAAUGGGCAGAG ...............................((((((((....(((.((((......)))))))((((((..((.....)).))))))..))))))))................... (-19.97 = -19.82 + -0.15)

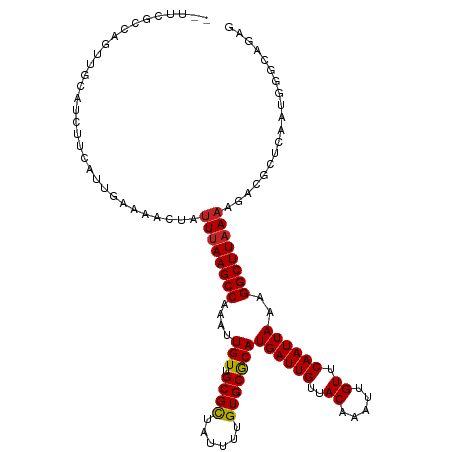
| Location | 20,543,509 – 20,543,624 |
|---|---|
| Length | 115 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Shannon entropy | 0.34212 |
| G+C content | 0.36036 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -18.88 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960776 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 20543509 115 - 24543557 CUCUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGCGAA-- ...(((((((((((..(..((((((((...(((((.((.....))........(((((........))))).))))).))))))))..)..))))))).((......))))))..-- ( -30.90, z-score = -2.61, R) >droSim1.chr3L 19895263 115 - 22553184 CUCUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGCGAA-- ...(((((((((((..(..((((((((...(((((.((.....))........(((((........))))).))))).))))))))..)..))))))).((......))))))..-- ( -30.90, z-score = -2.61, R) >droSec1.super_11 433894 115 - 2888827 CUAUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGCGAA-- ...(((((((((((..(..((((((((...(((((.((.....))........(((((........))))).))))).))))))))..)..))))))).((......))))))..-- ( -31.10, z-score = -2.74, R) >droYak2.chr3L 4185931 115 + 24197627 CUCUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGCGGG-- ..((((((((((((..(..((((((((...(((((.((.....))........(((((........))))).))))).))))))))..)..))))))).((......))))))).-- ( -34.00, z-score = -3.25, R) >droEre2.scaffold_4784 20237476 115 - 25762168 CUCUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGCGAA-- ...(((((((((((..(..((((((((...(((((.((.....))........(((((........))))).))))).))))))))..)..))))))).((......))))))..-- ( -30.90, z-score = -2.61, R) >droAna3.scaffold_13337 10896738 115 - 23293914 CUCUGGCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGACGCAACUGGCCAA-- ...((((((....((((((((((((((...(((((.((.....))........(((((........))))).))))).)))))).............))))))))...)))))).-- ( -35.51, z-score = -4.14, R) >dp4.chrXR_group8 7949421 107 - 9212921 ----------CUCUCCUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGGAGCUGCAACUGGCUGCAA ----------.........((((((((...(((((.((.....))........(((((........))))).))))).))))))))..........((.((((......)))).)). ( -24.50, z-score = -1.47, R) >droPer1.super_40 138919 107 - 810552 ----------CUCUCCUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGGAGCUGCAACUGGCUGCAA ----------.........((((((((...(((((.((.....))........(((((........))))).))))).))))))))..........((.((((......)))).)). ( -24.50, z-score = -1.47, R) >droWil1.scaffold_180949 4158078 117 + 6375548 UUCCUCUCUUUUUUUUUUCGUAAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCCAACGUUGGCUA ..............(((((((.(((((...(((((.((.....))........(((((........))))).))))).))))).....(....).))))))).((((....)))).. ( -23.60, z-score = -1.45, R) >droGri2.scaffold_15110 19274813 97 - 24565398 ----------------UUUUUUAAGCCUGUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAACGCAACAAUUUGGCUUAAAUAGUUUUCAAUGCAGACGAAAGCGUAG---- ----------------...((((((((...(((((.....(((((.(((....)))..))))).........))))).))))))))..((((((.........))))))....---- ( -17.84, z-score = -0.64, R) >droVir3.scaffold_13049 18803745 97 - 25233164 ----------------CUUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGCAGACGAAAGCGCAA---- ----------------((.((((((((...(((((.((.....))........(((((........))))).))))).)))))))).)).......(((.(.(....))))).---- ( -22.70, z-score = -2.02, R) >consensus CUCUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGCGAA__ ...................((((((((...(((((.((.....))........(((((........))))).))))).))))))))..((((((...)))))).............. (-18.88 = -18.95 + 0.08)
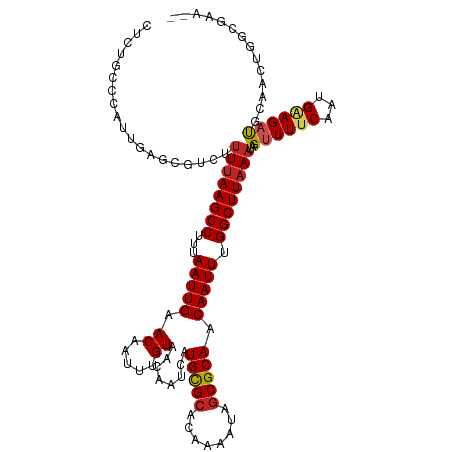
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:11 2011