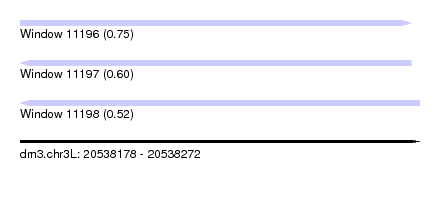
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 20,538,178 – 20,538,272 |
| Length | 94 |
| Max. P | 0.751804 |

| Location | 20,538,178 – 20,538,270 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 66.65 |
| Shannon entropy | 0.63473 |
| G+C content | 0.60480 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -13.55 |
| Energy contribution | -13.38 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.751804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20538178 92 + 24543557 -CUGCU--UUGGCUGUCGGACCGACUGCUGCAUCAUCUAACGCAGCAGCGGCA------GCUGCCAAGAUCGAUCGUCCUC-CUCCCGAUCCCUGCUGCAGC -(((((--(((((.((..(.(((.(((((((..........))))))))))).------)).))))))...(((((.....-....)))))......)))). ( -34.10, z-score = -1.68, R) >droEre2.scaffold_4784 20232482 89 + 25762168 -CUGCU--CUGGCUGCUGGAGCGACUGCUGCAUCAUCUAACGCAGCGGCGG---------CUGCCAAGAUCGAUCGUCCGC-CGCCCGAUCCCUGCUGCUCC -..(((--(..(...)..))))(.(((((((..........))))))))((---------(.((...(((((..((.....-))..)))))...)).))).. ( -30.70, z-score = 0.01, R) >droYak2.chr3L 4180500 89 - 24197627 -CUGCU--UUGGCUGCUGGAGCGACUGCUGCAUCAUCUAACGCAGCGGCGG---------CUGCCAAGAUCGAUCGUCCUC-CUCCCGAUGCCUGCUGCUCC -..((.--..(((....((((.(((.(((((..........)))))(((..---------..)))..........))))))-).......))).))...... ( -28.00, z-score = 0.23, R) >droSec1.super_11 428801 92 + 2888827 -CUGCU--UUGGCUGCCGGACCGACUGCUGCAUCAUCUAACGCAGCGGCGGCG------GCUGCCAAGAUCGAUCGUCCUC-CUCCCGAUCCCUGCUGCUGC -..(((--(((((.((((..(((.(((((((..........))))))))))))------)).))))))...(((((.....-....)))))...))...... ( -36.60, z-score = -2.14, R) >droSim1.chr3L 19890253 92 + 22553184 -CUGCU--UUGGCUGCCGGACCGACUGCUGCAUCAUCUAACGCAGCGGCGGCG------GCUGCCAAGAUCGAUCGUCCUC-CUCCCGAUCCCUGCUGCUGC -..(((--(((((.((((..(((.(((((((..........))))))))))))------)).))))))...(((((.....-....)))))...))...... ( -36.60, z-score = -2.14, R) >dp4.chrXR_group8 7942025 69 + 9212921 ---------------UUUGGAUGGCUGCUGCAUCAUCUAACGCAGCGGCGG---------CUGCCAAGAUCGAUCGUCCGC-UGCCUUUUGCCU-------- ---------------.(((((((((....))..))))))).(((((((((.---------.((.......))..)).))))-))).........-------- ( -23.40, z-score = -0.49, R) >droPer1.super_40 131926 69 + 810552 ---------------UUUGGAUGGCUGCUGCAUCAUCUAACGCAGCGGCGG---------CUGCCAAGAUCGAUCGUCCGC-UGCCUUUUGCCU-------- ---------------.(((((((((....))..))))))).(((((((((.---------.((.......))..)).))))-))).........-------- ( -23.40, z-score = -0.49, R) >droAna3.scaffold_13337 10891874 79 + 23293914 CUGGCUGACUAGCGGACUGGGCGACUGCUGCAUCAUCUAACGCAGCGGCGG---------CUGCCAAGAUCGAUCGUCCUC-UGACCGA------------- ((((....))))(((.(.(((((((((((((..........)))))))(((---------((....)).))).))))))..-.).))).------------- ( -26.00, z-score = -0.10, R) >droVir3.scaffold_13049 6423358 79 - 25233164 ---------------UCUCUGC--CUGCUGCAUCAUCUAACGCAGCGACGGCG------ACUGUUAAGAUCGAUCGUCGUUGUAUUUGACCGUUGCUGCUGA ---------------.......--..((.(((...((.((..(((((((((((------(((....)).))).)))))))))..)).))....))).))... ( -25.70, z-score = -1.71, R) >droMoj3.scaffold_6680 12415970 84 + 24764193 ---UGGCUGCGGCUGUGACUGUGGCUGCUGCAUCACCUAACGCAGCGACGGCG------ACUGUUAAGAUCGAUCGUCGUUGUGUUUGACCGU--------- ---..((.(((((..(....)..))))).)).(((...(((((((((((((((------(((....)).))).))))))))))))))))....--------- ( -36.70, z-score = -2.92, R) >droGri2.scaffold_15110 5284755 90 - 24565398 ---------CCACUGCCACAGU--CU-CUGCCUCAUCUAAUGCAGCGACGGCGGCGGUGACUGUUAAGAUCGAUCGUCGUUGUGUUUGACCGUUGCUGCUGC ---------...........((--(.-((((..........)))).))).(((((((..((.((((((..((((....))))..)))))).))..))))))) ( -30.80, z-score = -1.08, R) >consensus ___GCU___UGGCUGCCGGAGCGACUGCUGCAUCAUCUAACGCAGCGGCGGC________CUGCCAAGAUCGAUCGUCCUC_UGCCCGAUCCCUGCUGCUGC ......................(((.(((((..........)))))(((((.........)))))..........)))........................ (-13.55 = -13.38 + -0.17)

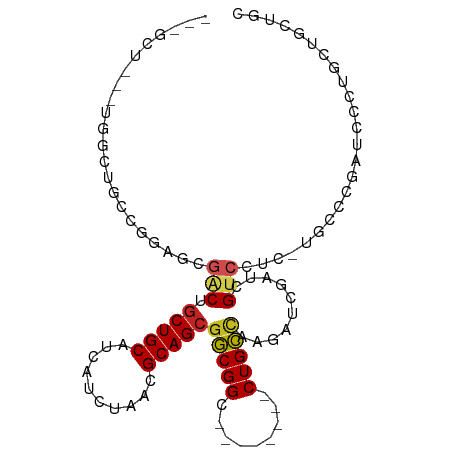
| Location | 20,538,178 – 20,538,270 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 66.65 |
| Shannon entropy | 0.63473 |
| G+C content | 0.60480 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -11.14 |
| Energy contribution | -11.15 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 20538178 92 - 24543557 GCUGCAGCAGGGAUCGGGAG-GAGGACGAUCGAUCUUGGCAGC------UGCCGCUGCUGCGUUAGAUGAUGCAGCAGUCGGUCCGACAGCCAA--AGCAG- .((((.((..(((((((..(-.....)..)))))))..)).((------(((((((((((((((....)))))))))).))).....))))...--.))))- ( -43.40, z-score = -2.88, R) >droEre2.scaffold_4784 20232482 89 - 25762168 GGAGCAGCAGGGAUCGGGCG-GCGGACGAUCGAUCUUGGCAG---------CCGCCGCUGCGUUAGAUGAUGCAGCAGUCGCUCCAGCAGCCAG--AGCAG- (((((.((.(((((((..((-.....))..)))))))(((..---------..)))((((((((....)))))))).)).))))).((......--.))..- ( -39.90, z-score = -1.64, R) >droYak2.chr3L 4180500 89 + 24197627 GGAGCAGCAGGCAUCGGGAG-GAGGACGAUCGAUCUUGGCAG---------CCGCCGCUGCGUUAGAUGAUGCAGCAGUCGCUCCAGCAGCCAA--AGCAG- (((((.(((((.(((((..(-.....)..))))))))(((..---------..)))((((((((....)))))))).)).))))).((......--.))..- ( -34.60, z-score = -1.03, R) >droSec1.super_11 428801 92 - 2888827 GCAGCAGCAGGGAUCGGGAG-GAGGACGAUCGAUCUUGGCAGC------CGCCGCCGCUGCGUUAGAUGAUGCAGCAGUCGGUCCGGCAGCCAA--AGCAG- ......((.((((((((..(-.....)..))))))))(((.((------((((((.((((((((....)))))))).).)))..)))).)))..--.))..- ( -41.00, z-score = -2.19, R) >droSim1.chr3L 19890253 92 - 22553184 GCAGCAGCAGGGAUCGGGAG-GAGGACGAUCGAUCUUGGCAGC------CGCCGCCGCUGCGUUAGAUGAUGCAGCAGUCGGUCCGGCAGCCAA--AGCAG- ......((.((((((((..(-.....)..))))))))(((.((------((((((.((((((((....)))))))).).)))..)))).)))..--.))..- ( -41.00, z-score = -2.19, R) >dp4.chrXR_group8 7942025 69 - 9212921 --------AGGCAAAAGGCA-GCGGACGAUCGAUCUUGGCAG---------CCGCCGCUGCGUUAGAUGAUGCAGCAGCCAUCCAAA--------------- --------.(((....(((.-((...(((......)))...)---------).)))((((((((....)))))))).))).......--------------- ( -27.30, z-score = -1.90, R) >droPer1.super_40 131926 69 - 810552 --------AGGCAAAAGGCA-GCGGACGAUCGAUCUUGGCAG---------CCGCCGCUGCGUUAGAUGAUGCAGCAGCCAUCCAAA--------------- --------.(((....(((.-((...(((......)))...)---------).)))((((((((....)))))))).))).......--------------- ( -27.30, z-score = -1.90, R) >droAna3.scaffold_13337 10891874 79 - 23293914 -------------UCGGUCA-GAGGACGAUCGAUCUUGGCAG---------CCGCCGCUGCGUUAGAUGAUGCAGCAGUCGCCCAGUCCGCUAGUCAGCCAG -------------..((..(-(.((((...((((...(((..---------..)))((((((((....)))))))).))))....)))).))..))...... ( -29.50, z-score = -1.49, R) >droVir3.scaffold_13049 6423358 79 + 25233164 UCAGCAGCAACGGUCAAAUACAACGACGAUCGAUCUUAACAGU------CGCCGUCGCUGCGUUAGAUGAUGCAGCAG--GCAGAGA--------------- ............(((.........((((..((((.......))------)).))))((((((((....)))))))).)--)).....--------------- ( -22.30, z-score = -0.75, R) >droMoj3.scaffold_6680 12415970 84 - 24764193 ---------ACGGUCAAACACAACGACGAUCGAUCUUAACAGU------CGCCGUCGCUGCGUUAGGUGAUGCAGCAGCCACAGUCACAGCCGCAGCCA--- ---------.((((...((.....((((..((((.......))------)).))))((((((((....)))))))).......))....))))......--- ( -23.80, z-score = -0.47, R) >droGri2.scaffold_15110 5284755 90 + 24565398 GCAGCAGCAACGGUCAAACACAACGACGAUCGAUCUUAACAGUCACCGCCGCCGUCGCUGCAUUAGAUGAGGCAG-AG--ACUGUGGCAGUGG--------- ((....))..(((((............)))))...........(((.(((((.(((.((((.((....)).))))-.)--)).))))).))).--------- ( -28.00, z-score = -0.85, R) >consensus GCAGCAGCAGGGAUCGGGCA_GAGGACGAUCGAUCUUGGCAG________GCCGCCGCUGCGUUAGAUGAUGCAGCAGUCGCUCCGACAGCCA___AGC___ .....................................................((.((((((((....)))))))).))....................... (-11.14 = -11.15 + 0.02)
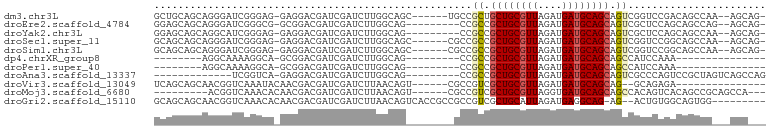

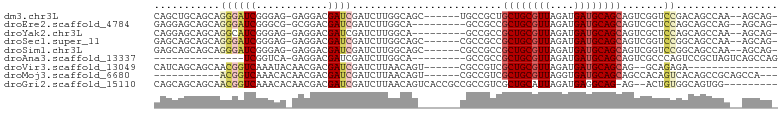
| Location | 20,538,178 – 20,538,272 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 68.86 |
| Shannon entropy | 0.60525 |
| G+C content | 0.60845 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -12.31 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
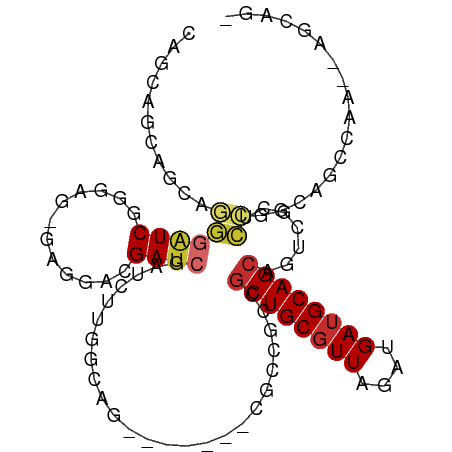
>dm3.chr3L 20538178 94 - 24543557 CAGCUGCAGCAGGGAUCGGGAG-GAGGACGAUCGAUCUUGGCAGC------UGCCGCUGCUGCGUUAGAUGAUGCAGCAGUCGGUCCGACAGCCAA--AGCAG- (((((((....((((((((..(-.....)..)))))))).)))))------))(((((((((((((....)))))))))).))).......((...--.))..- ( -44.20, z-score = -3.08, R) >droEre2.scaffold_4784 20232482 91 - 25762168 GAGGAGCAGCAGGGAUCGGGCG-GCGGACGAUCGAUCUUGGCA---------GCCGCCGCUGCGUUAGAUGAUGCAGCAGUCGCUCCAGCAGCCAG--AGCAG- ..(((((.((.(((((((..((-.....))..)))))))(((.---------...)))((((((((....)))))))).)).))))).((......--.))..- ( -40.10, z-score = -1.57, R) >droYak2.chr3L 4180500 91 + 24197627 CAGGAGCAGCAGGCAUCGGGAG-GAGGACGAUCGAUCUUGGCA---------GCCGCCGCUGCGUUAGAUGAUGCAGCAGUCGCUCCAGCAGCCAA--AGCAG- ..(((((.(((((.(((((..(-.....)..))))))))(((.---------...)))((((((((....)))))))).)).))))).((......--.))..- ( -34.80, z-score = -1.08, R) >droSec1.super_11 428801 94 - 2888827 GAGCAGCAGCAGGGAUCGGGAG-GAGGACGAUCGAUCUUGGCAGC------CGCCGCCGCUGCGUUAGAUGAUGCAGCAGUCGGUCCGGCAGCCAA--AGCAG- ........((.((((((((..(-.....)..))))))))(((.((------((((((.((((((((....)))))))).).)))..)))).)))..--.))..- ( -41.00, z-score = -2.13, R) >droSim1.chr3L 19890253 94 - 22553184 GAGCAGCAGCAGGGAUCGGGAG-GAGGACGAUCGAUCUUGGCAGC------CGCCGCCGCUGCGUUAGAUGAUGCAGCAGUCGGUCCGGCAGCCAA--AGCAG- ........((.((((((((..(-.....)..))))))))(((.((------((((((.((((((((....)))))))).).)))..)))).)))..--.))..- ( -41.00, z-score = -2.13, R) >droAna3.scaffold_13337 10891874 79 - 23293914 ---------------UCGGUCA-GAGGACGAUCGAUCUUGGCA---------GCCGCCGCUGCGUUAGAUGAUGCAGCAGUCGCCCAGUCCGCUAGUCAGCCAG ---------------..((..(-(.((((...((((...(((.---------...)))((((((((....)))))))).))))....)))).))..))...... ( -29.50, z-score = -1.49, R) >droVir3.scaffold_13049 6423358 81 + 25233164 CAUCAGCAGCAACGGUCAAAUACAACGACGAUCGAUCUUAACAGU------CGCCGUCGCUGCGUUAGAUGAUGCAGCAG--GCAGAGA--------------- ..............(((.........((((..((((.......))------)).))))((((((((....)))))))).)--)).....--------------- ( -22.30, z-score = -0.72, R) >droMoj3.scaffold_6680 12415970 84 - 24764193 -----------ACGGUCAAACACAACGACGAUCGAUCUUAACAGU------CGCCGUCGCUGCGUUAGGUGAUGCAGCAGCCACAGUCACAGCCGCAGCCA--- -----------.((((...((.....((((..((((.......))------)).))))((((((((....)))))))).......))....))))......--- ( -23.80, z-score = -0.47, R) >droGri2.scaffold_15110 5284755 92 + 24565398 CAGCAGCAGCAACGGUCAAACACAACGACGAUCGAUCUUAACAGUCACCGCCGCCGUCGCUGCAUUAGAUGAGGCAG-AG--ACUGUGGCAGUGG--------- ..((....))..(((((............)))))...........(((.(((((.(((.((((.((....)).))))-.)--)).))))).))).--------- ( -28.20, z-score = -0.86, R) >consensus CAGCAGCAGCAGGGAUCGGGAG_GAGGACGAUCGAUCUUGGCAG_______CGCCGCCGCUGCGUUAGAUGAUGCAGCAGUCGCUCCGGCAGCCAA__AGCAG_ ...........((((((............)))).........................((((((((....)))))))).......))................. (-12.31 = -12.40 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:40:09 2011